Introduction
Bombyx mori is a female heterogametic lepidopteran in which the sex chromosomes are designated ZW for the female and ZZ for the male. Sex in silkworm is controlled by the presence/absence of the W chromosome (Hashimoto, Reference Hashimoto1933). The W chromosome is heterochromatic and composed largely of repeated sequences, with little evidence for the presence of expressed genes except on autosomal translocations (Abe et al., Reference Abe, Seki, Ohbayashi, Tanaka, Yamashita, Fujii, Yokoyama, Takahashi, Banno, Sahara, Yoshido, Ihara, Yasukochi, Mita, Ajimura, Suzuki, Oshiki and Shimada2005). By contrast, 17 visible mutations have been localized to the Z chromosome, including the ones affecting larval cuticle transparency (os, sex-linked translucent, 1–0·0, and od, distinct translucent, 1–49·6), body shape (e, elongated, 1–36·4), molting (nm-s, non-molting-s, 1–3·0), maturity (Lm, late maturity, 1–2·0) and body colour (sch, sex-linked chocolate, 1–21·5), as well as several mutations affecting other developmental stages (Fujii et al., Reference Fujii, Banno, Doira and Kawaguchi1998). sch is a ‘classical’ marker expressed in newly hatched larvae and used to facilitate early-stage sexing for silk production. Male silkworms produce a higher ratio of silk per unit food consumed than females (Zhu et al., Reference Zhu, Chen, Zhao, Lu and Xiang2001); hence, identification of molecular markers close to sch or positional cloning of the gene itself could lead to the development of tools for marker-assisted selection. Further, construction of a detailed molecular linkage map for the Z chromosome will enable cloning of sch and other mutations of interest.
Several groups have constructed molecular linkage maps in the silkworm by using a variety of molecular markers. They include a high-density randomly amplified polymorphic DNA (RAPD) map with 1018 markers (Yasukochi, Reference Yasukochi1998; Yasukochi et al., Reference Yasukochi, Ashakumary, Baba, Yoshido and Sahara2006), a medium-density amplified fragment length polymorphism (AFLP) map with 356 markers (Tan et al., Reference Tan, Wan, Zhu, Lu, Xiang and Deng2001), and a low density restriction fragment length polymorphism (RFLP) map based on 189 maternal EST cDNA clones (Nguu et al., Reference Nguu, Kadono-Okuda, Mase, Kosegawa and Hara2005). In these maps, only Yasukochi et al. (Reference Yasukochi, Ashakumary, Baba, Yoshido and Sahara2006), Tan et al. (Reference Tan, Wan, Zhu, Lu, Xiang and Deng2001) and Prasad et al. (Reference Prasad, Muthulakshmi, Madhu, Archak, Mita and Nagaraju2005) developed a significant number of markers on the Z chromosome; they did this by using the sex of the insect or the sex-linked mutation, od. Maps based on RAPDs and AFLPs are useful only for the strains in which they were constructed and the results cannot be extrapolated to other strains (Kadono-Okuda et al., Reference Kadono-Okuda, Kosegawa, Mase and Hara2002).
Microsatellites or simple sequence repeats (SSRs) are tandemly repeated units of one to six nucleotides which are abundant in prokaryotic and eukaryotic genomes (Field & Wills, Reference Field and Wills1996). They are ubiquitously distributed in the genome, both in protein-coding and non-coding regions (Toth et al., Reference Toth, Gaspari and Jurka2000). The advent of PCR and the availability of high-throughput automated sequencers have increased the use of SSR markers, which have become an informative, widely used and versatile class of genetic markers (Litt & Luty, Reference Litt and Luty1989; Schlötterer, Reference Schlötterer2004). SSR techniques have proven to be convenient and reliable tools to generate highly polymorphic molecular markers that greatly facilitate building linkage maps. Once a linkage map is constructed in one laboratory, it can be readily shared with other laboratories. All that is required is the synthesis of polymorphic SSR primer pairs for the population under study. Nagaraja et al. (Reference Nagaraja, Mahesh, Satish, Madhu, Muthulakshmi and Nagaraju2005) constructed a linkage map for the Z chromosome by using mixed dominant and co-dominant molecular markers, including RAPDs and SSRs. However, in this Z map, the main markers are dominant RAPDs, and the sex-linked behaviour of the two SSR markers was not discussed.
We constructed an SSR linkage map for silkworm, which covered all 28 chromosomes of silkworm (Miao et al., Reference Miao, Xu, Li, Li, Huang, Dai, Marino, Mills, Zeng, Mita, Jia, Zhang, Liu, Xiang, Guo, Xu, Kong, Lin, Shi, Lu, Zhang, Huang, Yasukochi, Sugasaki, Shimada, Nagaraju, Xiang, Wang, Goldsmith, Lu, Zhao and Huang2005). During linkage analysis, we found that some SSR loci segregated in disagreement with the expected Mendelian genetic ratio of 1:1 in a backcross population. In the beginning, we considered to discard these markers. We further investigated these loci to determine the cause of their unexpected segregation pattern. As a result, we discovered that they were sex-linked. Here, we analyse the genetic character of such co-dominant sex-linked SSR markers, construct a linkage map for the Z chromosome, and map the location of the visible mutation sex-linked chocolate (sch).
Materials and methods
Silkworm strains and genetic crosses
Two silkworm strains K05 (homozygous for sch ) and C108 (wild-type for sch) were obtained from the Sericultural Research Institute, Chinese Academy of Agricultural Sciences (Zhenjiang, People's Republic of China). A single-pair mating of a female C108 crossed with a male K05 was performed to obtain the F1 population. Ten F1 individuals (five females and five males) were selected for SSR marker linkage analysis. A single F1 male was backcrossed with a K05 female to produce a BC1M population. A total of 188 BC1M individuals were analysed: 94 sch segregants and 94 wild-type segregants.
DNA extraction and SSR linkage analysis
Genomic DNA was prepared following the method of Yasukochi (Reference Yasukochi1998). SSR locus screening and analysis was performed using the methods of Miao et al. (Reference Miao, Xu, Li, Li, Huang, Dai, Marino, Mills, Zeng, Mita, Jia, Zhang, Liu, Xiang, Guo, Xu, Kong, Lin, Shi, Lu, Zhang, Huang, Yasukochi, Sugasaki, Shimada, Nagaraju, Xiang, Wang, Goldsmith, Lu, Zhao and Huang2005). In the present study, to confirm whether the SSR markers belonged to the Z chromosome, we selected ten F1 individuals from the C108×K05 cross. If a marker belongs to the Z chromosome, all F1 females (ZW) will have a single band, whereas all the F1 males (ZZ) will have two bands. Using these criteria, we found one PCR marker and six SSR markers were located on the Z chromosome. As a contrast, we chose one autosomal SSR marker (S1126) to confirm our earlier results (Miao et al., Reference Miao, Xu, Li, Li, Huang, Dai, Marino, Mills, Zeng, Mita, Jia, Zhang, Liu, Xiang, Guo, Xu, Kong, Lin, Shi, Lu, Zhang, Huang, Yasukochi, Sugasaki, Shimada, Nagaraju, Xiang, Wang, Goldsmith, Lu, Zhao and Huang2005), i.e. all the autosomal SSR markers in the ten F1 individuals should have two bands, regardless of sex.
Linkage analysis and map construction
A Z chromosome genetic linkage map was constructed based on the segregation of polymorphic SSR markers and sch phenotypes in 188 F1 male backcross (BC1M) offspring. The linkage analysis and map construction were carried out using Mapmaker 3.0 software (Lander et al., Reference Lander, Green, Abrahamson, Barlow and Daly1987).
Results
Co-dominant SSR markers in BC1M segregation
Figure 1 shows the data for an SSR marker in a BC1M population. Figure 1 a shows the patterns for sch individuals (sch/sch), whereas Fig. 1 b shows the patterns for wild-type individuals (+sch/sch). Both the mutant sch phenotype and its wild-type are found with three kinds of SSR band patterns: two homozygous single bands (designated M1 or M2) and one heterozygous double band designated M1M2.

Fig. 1. Representative results for a co-dominant SSR marker in the BC1M population K05×(C108×K05). (a) All individuals have the phenotype of the visible mutation sch. (b) All individuals are wild-type (+sch). M1 and M2 are two kinds of homozygotes with single SSR bands and M1M2 are heterozygotes with two bands. Based on the model shown in Fig. 2, because all individuals in (a) are phenotypically mutant (sch), only those expressing M2 bands are the parental type, whereas all individuals expressing M1 and M1M2 bands are recombinant type; by contrast, in (b), all those with M2 bands are recombinant type, and all M1 and M1M2 bands are parental type.
Analysis and identification of co-dominant SSR markers on the Z chromosome or on an autosome
When an SSR marker is linked to a recessive visible marker, SSR bands will differ if the co-dominant marker is located on the Z chromosome or on an autosome. In Fig. 2 and Table 1, we diagram a model of the inheritance pattern of a visible mutation and a linked co-dominant SSR marker in a backcross population between an F1 male and a homozygous mutant parental female. We suppose that M is a polymorphic SSR marker between the C108 and K05 parents. Because no crossover occurs in silkworm females (Sturtevant, Reference Sturtevant1915; Goldsmith, Reference Goldsmith, Goldsmith and Willkins1995), they produce only two kinds of gametes. Table 1 shows the Mendelian ratios expected for two linked markers in F1 and BC1M segregants (see the ‘Parental type’ column). If the SSR marker is located on the Z chromosome, in the F1 generation, two genotypes and two kinds of SSR band patterns are produced: two bands (M1M2) and one band (M2). If the SSR marker is located on an autosome, there is only one genotype and only one SSR band pattern will be produced (M1M2; Table 1 and Fig. 2).

Fig. 2. Inheritance models for a visible mutation on the Z chromosome with a linked polymorphic SSR marker in a backcross population with complete linkage in the female (BCF). ‘M’ refers to the microsatellite locus; M1 and M2 refer to the polymorphism between the two parents.
Table 1. The genotype and the phenotype of an SSR marker (MM) linked to a recessive visible marker (schsch) on the Z chromosome or on an autosome in BC1M segregantsFootnote a

a The two parental genotypes are C108: M1+sch/M1+sch and K05: M2sch/M2sch, respectively.
In Table 1 and Fig. 2 of the ‘parental type’, if we designate the homozygote bands (M1 and M2) as 1 and heterozygote bands (M1M2) as 2, we predict a 3:1 ratio in the BC1M for markers segregating on the sex chromosome. The ‘3’ comes from all of the females and half of the males, which express a single band (3/4); the ‘1’ comes from the other half of male individuals, which display a heterozygous double banding pattern (1/4). This differs from the 1:1 inheritance pattern of SSR markers predicted on autosomes (Table 1). In contrast, the phenotypic ratio for the sex-linked sch mutation is 1:1 for both autosomes and the Z chromosome. So, in Fig. 1 a, all M2 bands are parental type; in Fig. 1 b, all M2 bands are recombinant type.
Figure 3 shows the band patterns of co-dominant SSR markers in the F1 generation. S0105 is located on the Z chromosome, so F1 females have one band (M2) and F1 males have two bands (M1M2) (Fig. 3 a). A total of six SSR markers with this heterozygous/homozygous structure were identified. S1126 is located on an autosome, so both F1 females and males have two bands (M1M2) (Fig. 3 b).

Fig. 3. (a) The band patterns of a Z chromosome SSR marker (S0105) in F1 female and male individuals. (b) The band patterns of an autosomal SSR marker (S1126) in F1 female and male individuals. Lanes 1–5 are F1 females; lanes 6–10 are F1 males. The inner ladders are internal size markers.
Statistical results for the BC1M population are shown in Table 2. The genotypic ratio of single SSR bands (M1+M2) to double bands (M1M2) did not differ significantly from the predicted ratio (Table 2) of 3:1 for the four SSR markers tested. The phenotypic ratio of sch to +sch did not differ from the predicted ratio (Table1) of 1:1. These results are consistent with the model of sex-linked inheritance in Table 1 and Fig. 2.
Table 2. SSR band patterns in sch and +sch segregants of K05×(C108×K05)
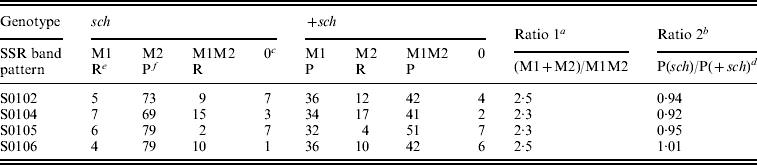
a Ratios do not differ significantly from the predicted ratio of 3:1 (chi-squared test, P>0·05).
b Ratios do not differ significantly from the predicted ratio of 1:1 (chi-squared test, P>0·05).
c The number 0 indicates that these individuals were not amplified by PCR.
d In the +sch segregants, the P type includes M1 and M1M2 (see Table 1).
e ‘R’ indicates the recombinant type.
f ‘P’ indicates the parental type.
Adjustment of SSR band patterns in the BC1M population for linkage mapping
As shown in Fig. 1, both the mutant sch phenotype and its wild-type are found with three kinds of SSR band patterns. It is difficult to construct a linkage map using Mapmaker software in such a backcross model.
To resolve this problem, according to the inheritance model of Fig. 2, we have to adjust the banding pattern to fit a backcross model. In the sch segregants (Fig. 1 a), the M2 single bands were read as 1 to indicate that they were parental type, and the single band M1 and the double bands M1M2 were recoded as 2 to indicate that they were recombinant types. By contrast, in the wild-type segregants (Fig. 2 b), the single band M2 was coded as 1, to indicate that they were recombinant type, while the single band M1 plus the double bands M1M2 were coded as 2 for parental types. Using these coded data, we constructed a Z chromosome linkage map using Mapmaker software.
Construction of Z chromosome genetic map
Between the parents C108 and K05, we found one PCR marker and six SSR markers that had Z chromosome-linked polymorphic character. When data were coded as above, all the six SSR polymorphic markers segregated in the expected ratio of 1:1 (P>0·05, chi-squared test, df=1). At a logarithm of odds (LOD) score cut-off of 3·0, using the ‘group’ command of Mapmaker software, the six SSR markers, one PCR marker and the visible mutation sch aggregated in one group; using the ‘order’ and ‘map’ commands, we derived a Z chromosome linkage map with the sch gene located between the S0105 and S0106 SSR loci (Fig. 4). The map covers 76·1 cM.

Fig. 4. Z chromosome linkage map. The visible mutation sch was localized by analysing the BCM backcross population of K05×(C108×K05).
Discussion
In the domesticated silkworm, the Z chromosome is very important not only in evolution but also in economics. Constructing a fine structure linkage map for the Z chromosome is of benefit for map-based cloning and for marker-assisted selection. Tan et al. (Reference Tan, Wan, Zhu, Lu, Xiang and Deng2001) and Prasad et al. (Reference Prasad, Muthulakshmi, Madhu, Archak, Mita and Nagaraju2005) mapped the Z chromosome using sex or the sex-linked gene od and RAPD or AFLP molecular markers. We know that RAPDs and AFLPs are dominant molecular markers that only indicate the presence or absence of a band for a specific allele. Co-dominant molecular markers can provide more information, because two or more alleles can segregate visibly. Nagaraja et al. (Reference Nagaraja, Mahesh, Satish, Madhu, Muthulakshmi and Nagaraju2005) constructed a Z chromosome linkage map by mixed markers, which involved two SSR markers. We have tried to combine this map with our SSR map; unfortunately, there was no polymorphism in our mapping population for the two SSR markers reported in this paper.
When we constructed a linkage map for silkworm using co-dominant SSR markers, some loci did not show the expected 1:1 Mendelian inheritance pattern in backcross segregation (Miao et al., Reference Miao, Xu, Li, Li, Huang, Dai, Marino, Mills, Zeng, Mita, Jia, Zhang, Liu, Xiang, Guo, Xu, Kong, Lin, Shi, Lu, Zhang, Huang, Yasukochi, Sugasaki, Shimada, Nagaraju, Xiang, Wang, Goldsmith, Lu, Zhao and Huang2005). Instead, they had a ‘pseudo-ratio’ of 3:1 of single band/double band, because there were two kinds of homozygotes producing single bands (M1 and M2) as well as a double-banded heterozygote (M1M2; Fig. 1). According to the inheritance model of a co-dominant SSR marker together with a linked visible mutation, we can distinguish the parental and recombinant types to determine the frequency of recombinants, providing an efficacious method to detect linkage to the Z chromosome by the presence of an aberrant 3:1 ratio. Subsequently, we recoded the data to take advantage of the capabilities of Mapmaker software and generate an accurate linkage map for co-dominant markers.
As part of our study, we mapped the sch mutation, which is widely used for male silkworm selection in silkworm breeding in China. Future research will focus on finding more closely linked molecular markers to facilitate marker-assisted selection with a long-term goal of cloning the sex-linked chocolate gene.
This work was supported by the Natural Science Foundation of China (30471310). We sincerely thank Dr Sally C. Levings, Professor Leonard Rabinow and Professor Marian R. Goldsmith for their comments and revision of the draft of this paper.








