1. Introduction
Sex determination in the housefly, Musca domestica, is more variable than in most other species, which usually exhibit just a single sex-determining mechanism (Bull, Reference Bull1983; Dübendorfer et al., Reference Dübendorfer, Hediger, Burghardt and Bopp2002). Polymorphism for sex-determining factors has been found in many natural populations of the housefly (Franco et al., Reference Franco, Rubini and Vecchi1982; Denholm et al., Reference Denholm, Franco, Rubini and Vecchi1985; Tomita & Wada, Reference Tomita and Wada1989b; Feldmeyer et al., submitted; Table 1). In ‘standard’ strains, sex is determined by a male-determining factor, M, which is located on the Y chromosome; therefore males are XY and females are XX. During development, the M factor blocks the female-determining factor F located on autosome IV, the activity of which is necessary for female development. In many populations, M is located on one of the autosomes or even on the X chromosome (Denholm et al., Reference Denholm, Franco, Rubini and Vecchi1983). In such populations, usually a dominant constitutive mutation of F (F D) is also present, which triggers female development even in the presence of several M factors in the same individual (see McDonald et al., Reference McDonald, Evenson, Nickel and Johnson1978; Franco et al., Reference Franco, Rubini and Vecchi1982; Dübendorfer et al., Reference Dübendorfer, Hediger, Burghardt and Bopp2002; Table 1).
Table 1. Relation between genotype and gender in the housefly
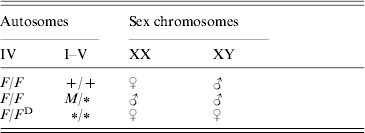
The female-determining factors (F/F D) are located on autosome IV. The male-determining factors (M) can be located on any chromosome. A ‘+’ indicates the wild-type state (no M) and a ‘∗’ indicates that an M or+allele on this locus will not influence the sex.
The XY system is probably ancestral in the housefly, since it is also very common in closely related species (Boyes et al., Reference Boyes, Paterson and Corey1964) and the first reports on autosomal sex-determining (SD) factors appeared only around 1960 (reviewed by Franco et al., Reference Franco, Rubini and Vecchi1982). Since then, the geographical distribution of different SD factors has been studied on most of the continents and appears to follow geographical clines. In general, the Y chromosome is more common at higher latitudes and altitudes and its frequency gradually decreases with decreasing latitude and altitude, leading to populations with only autosomal sex-determining factors (autosomal M and F D) closer to the equator and at low altitudes (Franco et al., Reference Franco, Rubini and Vecchi1982; Tomita & Wada, Reference Tomita and Wada1989b; Çakir & Kence, Reference Çakir and Kence1996; Hamm et al., Reference Hamm, Shono and Scott2005; Feldmeyer et al., submitted). It is not clear what forces are responsible for the distribution of different SD factors, but temperature seems to be an important factor (Feldmeyer et al., submitted).
There is some evidence that autosomal sex-determining factors have spread in some populations replacing the standard XY system (Franco et al., Reference Franco, Rubini and Vecchi1982; Tomita & Wada, Reference Tomita and Wada1989a, Reference Tomita and Wadab). It has been hypothesized (Franco et al., Reference Franco, Rubini and Vecchi1982; Denholm et al., Reference Denholm, Franco, Rubini and Vecchi1985; Tomita & Wada, Reference Tomita and Wada1989a, Reference Tomita and Wadab; Çakir & Kence, Reference Çakir and Kence1996) that the observed distributions are a transient state. In particular, Franco and colleagues (Reference Franco, Rubini and Vecchi1982) suggested that autosomal M factors are spreading north in Europe, but their hypothesis was based only on the change in frequency of the Y chromosome in a few populations before 1980. No systematic or recent studies have been done on the dynamics of different SD factors in natural populations of the housefly. The last study in continental Europe dates from 25 years ago (Franco et al., Reference Franco, Rubini and Vecchi1982), in which cytological data were used to show a clear latitudinal cline with the standard XY system exclusively present in the north of Europe (Iceland, Denmark, the Netherlands, Germany and Switzerland) and entirely autosomal populations (lacking the Y chromosome) in southern Italy at altitudes below 100 m. In northern Italy mixed populations have been found, with the frequency of the Y chromosome increasing with higher altitudes and latitudes.
The aim of this study was to investigate whether the distribution of SD factors in the housefly has changed in Europe over the last 25 years. Therefore, we sampled a number of European populations on a north–south transect from Germany to southern Italy, and compared the frequency of males that carry the Y chromosome and autosomal M factors with the data published by Franco and colleagues (Reference Franco, Rubini and Vecchi1982). Additionally, we analysed the frequencies of the F D factor and we publish the frequencies of M factors located on different chromosomes, which has not been done before for European housefly populations.
2. Materials and methods
(i) Collection and rearing of flies
We sampled populations along a north–south transect from north Germany to south Italy in July 2006 (see Fig. 1 and Table 2 for details on the sampling locations). Most of the sampling sites were chosen to be close to the ones studied by Franco et al. (Reference Franco, Rubini and Vecchi1982), as far as we could judge from the limited information available. For Germany and Switzerland, they gave only the name of a state (Baden-Württemberg) or a canton (Mittelland) and our sampling sites lie within these areas. For Italy, Franco and colleagues published a map indicating sampling sites together with information on altitudes, but precise geographical coordinates were lacking. We judged their locations visually and used altitudes within 110 m, but usually within a 50 m range. The exception is population IT5 where the altitude given by Franco et al. (Reference Franco, Rubini and Vecchi1982) does not match the area indicated by them, so to match the altitude we sampled 50 km west of their indicated location. Ultimately, our sampling sites were distributed approximately homogeneously along a north–south transect, with some areas having sampling sites at different altitudes.

Fig. 1. Sampling locations in the study of Franco and colleagues (Reference Franco, Rubini and Vecchi1982; dots) and in the present study (circles). Locations from the present study are labelled with population codes as in Table 2.
Table 2. Geographical coordinates, altitudes (in metres above sea level) and average yearly temperatures of the sampling sites

Sampling sites are ordered according to their latitude.
Letters in the code indicate the country of origin: GE, Germany; SW, Switzerland; IT, Italy.
For each location, we obtained data on average monthly minimum and maximum temperatures from WORLDCLIM (www.worldclim.org; see Hijmans et al., Reference Hijmans, Cameron, Parra, Jones and Jarvis2005), which provides global estimates at a resolution of 1 km2. We estimated average yearly temperatures as the mid-point between minimum and maximum temperatures (Table 2). Since all these measures of temperature are highly correlated (P<0·0001, Pearson's product-moment correlation test), we used only the average yearly temperature in our statistical analysis (see below).
Flies were sampled at farms and horse stables. At each location we caught approximately 50 adult males and females (except for IT3, where only 10 females were found). The flies were caught with sweeping nets, placed in plastic containers and provided with water and milk powder as food. They were also provided with egg-laying medium (according to Hilfiker-Kleiner et al., Reference Hilfiker-Kleiner, Dübendorfer, Hilfiker and Nöthiger1994) on which females laid eggs within a few days. Larvae were transferred to larger containers after a few days and fed ad libitum on the same medium. Flies from all the locations (or their offspring) were successfully transported to the laboratory and populations were established and maintained in cages at a population size of approximately 500 individuals.
(ii) Analysis of the sex-determining factors
(a) M factors
The presence of different M factors in males was determined by two generations of single-pair crosses with standard XX (without an F D factor) virgin females, from a marker strain that carries visible recessive mutations in the homozygous state on each of the five autosomes (Tomita & Wada, Reference Tomita and Wada1989b). The sex ratio of F1 offspring shows whether the father was homozygous for at least one M factor (only sons are produced) or heterozygous for all M factors (daughters are also present among the offspring). Sex-linked inheritance of visible markers in the second generation of backcrosses to marker-strain females shows on which chromosomes M factors are located. This is a standard procedure in our laboratory and it gives a good estimation of the frequency of M factors located on different autosomes (for details see Denholm et al., Reference Denholm, Franco, Rubini and Vecchi1983). However, if a focal wild-type male was homozygous for M (producing all-male offspring) and all his sons appeared to have two (or more) M factors (e.g. M on autosome II and V), we could not unambiguously determine whether the father was homozygous for M on only one or on both chromosomes, especially if the number of sons was small. For example, M II/M II; M V/M V, M II/+; M V/M V and M II/M II; M V/+ males all produce M II/+; M V/+ sons when mated with standard females. This happened a few times (13 males in total, with a maximum of 4 males per population). For each chromosome involved in a population, we calculated both the minimal frequency of M (assuming that all ambiguous males were heterozygous for M) and the maximal frequency of the M factor (assuming that all ambiguous males were homozygous for M) on the given chromosome. We then used the mid-point value between the two extremes as a population estimate.
We used 20 males from each population for the first series of crosses and 3 sons from each of them for the F1 backcrosses (although we did not obtain offspring from all males). Males used for analysis were either the ones caught in the field (IT3), or from the first generation in the laboratory (offspring of the wild-caught flies: IT6, IT7, IT8, IT10, IT11, IT12), the third generation in the laboratory (GE1, GE2, SW, IT1, IT2, IT4, IT5) or the fourth generation (IT9). Because of the lack of visible markers on the X and the Y chromosomes, in cases in which we assigned M to a sex chromosome, we cannot be sure whether it was located on the Y or the X (as has been found in Britain: Denholm et al., Reference Denholm, Franco, Rubini and Vecchi1983, Reference Denholm, Franco, Rubini and Vecchi1985). If M was located on a sex chromosome we will call this chromosome Y, but we will discuss this issue in more detail later.
(b) F D factor
F and F D factors have been sequenced at the University of Zürich (M. Hediger and D. Bopp, personal communication). F D has two deletions compared with F in all populations analysed (of European, Asian and African origin). We used primers designed for one of these deletions to distinguish between F (one band present) and F D (two bands) females. We used approximately 20 females from each population, either females caught in the field (populations GE2, SW, IT5 and all 10 females from IT3) or from the first generation in the laboratory (all the other populations). Additionally, we took 2 or 3 females from each population and crossed them individually with a male homozygous for M located on autosome III. Females without F D produce only sons, but those with F D also produce daughters, because F D is dominant over M. After determining the sex of the offspring, we also analysed the mothers molecularly and found without exception that the results of the molecular analysis were consistent with those obtained from the crosses. This shows that the deletion in the F D factor is also present in the populations we collected and justifies the use of the molecular technique for analysing frequencies of F D in our populations.
(iii) Statistical analysis
We performed a logistic regression analysis using the glm function with quasi-binomial errors in R (R Development Core Team, 2006) to investigate the influence of latitude, altitude and temperature on the frequency of autosomal M males (with at least one autosomal M factor) and on the frequency of females with the F D factor. We started with a full model (including all two-way interactions between explanatory variables) and used backward selection to find the minimal adequate model. The significance of the difference between models was assessed with the likelihood-ratio approach, using F-tests to correct for under- and overdispersion (Krackow & Tkadlec, Reference Krackow and Tkadlec2001).
A statistical comparison between the frequencies of different SD factors in the past and present is only possible to a limited extent, since Franco et al. (Reference Franco, Rubini and Vecchi1982) performed only cytological observations. They used the frequency of XX males as a measure for the frequency of autosomal males. They checked the linkage of autosomal M factors with crosses similar to ours, but they do not provide the exact frequencies of different factors. They also do not provide data on frequencies of the F D factor. Moreover, due to the lack of data on the number of males tested by Franco and colleagues (Reference Franco, Rubini and Vecchi1982), in each autosomal and standard population separately (except for GE2), we could only include eight populations (GE2, IT2, IT3, IT4, IT5, IT7, IT8 and IT10) in a statistical analysis to compare frequencies of autosomal males (without a Y chromosome) between our study and theirs. For this analysis, we performed a mixed-model logistic regression analysis in R using the lmer function with binomial errors from the lme4 package. The full model included population as a random effect and ‘study’ (Franco et al., Reference Franco, Rubini and Vecchi1982 or this study) as a fixed effect. Significance of the effect of ‘study’ was judged using the likelihood-ratio approach, using an F-test to correct for overdispersion (Krackow & Tkadlec, Reference Krackow and Tkadlec2001). For each of the eight populations we also performed a binomial test, to see whether there is a significant change in the frequency of XX males between the past and the present.
3. Results
(i) Distribution of sex-determining factors in 2006
We found M factors on the sex chromosomes and on each of the autosomes (Table 3, Fig. 2 a). M located on autosome III was the most frequent among autosomal M factors and the frequencies of M on autosomes IV and V were very low. We did not detect any autosomal M in the German and Swiss populations or in one northern Italian population from the highest altitude (IT3). In populations with autosomal SD factors, often single males with multiple M factors, located on up to four different chromosomes, were observed (data not shown). Statistical analysis showed that altitude, latitude, temperature and interaction of temperature and latitude (and to a lesser extent interaction between temperature and altitude) influence the frequencies of autosomal M males (Table 4).

Fig. 2. Distribution of sex-determining factors in the housefly in 2006. (a) Relative frequencies of M factors located on different chromosomes: white, sex chromosome; yellow, autosome I; red, autosome II; green, autosome III; blue, autosome IV; pink, autosome V. (b) Frequencies of females with (red) and without (blue) the F D factor.
Table 3. Estimated frequencies of females with F D factor and frequencies of M factors in males in samples from different housefly populations

Table 4. Logistic regression analysis of (a) frequencies of autosomal M males and (b) frequencies of females with F D

Parameter estimates (logit scale) and their standard errors (SE) are shown for the final models, after the removal of non-significant variables.
Temperature refers to the average yearly temperature.
Δdev indicates the change in deviance resulting from removing the given variable from the final model. The F-tests for significance of removed variables have 1 and residual degrees of freedom of the final model (DF) for numerator and denominator, respectively.
Final models: (a) deviance=3·05, residual DF=9; (b) deviance=27·28, residual DF=11.
We did not find F D in populations in Germany and only at low frequencies in Switzerland and at the highest location in northern Italy (IT3; Table 3, Fig. 2 b). In most of the Italian populations frequencies of F D females were above 0·75, and in three populations F D appeared to be at fixation. Statistical analysis showed that the frequency of females with F D is influenced by latitude, temperature and the interaction of the two (Table 4).
(ii) Comparison with the past
A comparison between our results and those of Franco and colleagues (Reference Franco, Rubini and Vecchi1982) shows that there is no clear evidence for the spread of autosomal M factors northwards during the last 25 years (Fig. 3). In the two northernmost populations and in IT3, which lacked XX males in the past, we also did not find any autosomal M factor. Furthermore, all populations described by Franco and colleagues (Reference Franco, Rubini and Vecchi1982) as mixed or autosomal were found to have autosomal M factors in 2006. However, in the populations which were described by Franco and colleagues as autosomal in 1982 (IT6, IT9 and IT12) we also found M on a sex chromosome. Statistical analysis based on the eight populations for which comparable data were available shows no significant systematic change in the frequencies of autosomal males in recent decades (Table 5). Statistical analysis for each population separately, shows a significant decrease in the frequency of XX males for two populations: IT5 and IT8 (P<0·002, which is also significant after Bonferroni correction for multiple tests).

Fig. 3. Comparison of karyotype frequencies in males in the past and the present (2006). For each population the left-hand bar corresponds to the data from Franco et al. (Reference Franco, Rubini and Vecchi1982) and the right-hand bar to the data from this study. We inferred karyotypes from our crosses assuming that Y is the sex chromosome bearing the M factor (see Section 2). Three populations analysed by us are not included in the figure since they were not studied by Franco and colleagues. Populations are ordered according to decreasing latitude of the sampling sites (see Table 2).
Table 5. Logistic mixed-model analysis of the frequencies of XX males in the study of Franco et al. (Reference Franco, Rubini and Vecchi1982) and this study
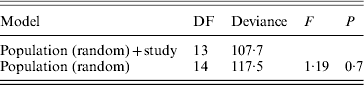
The full model includes population as a random effect and study (data from Franco et al., Reference Franco, Rubini and Vecchi1982 or from our study) as a fixed effect under analysis.
No significant difference between studies was found.
The distribution of F D also seems to be relatively stable over time. F D frequencies were not analysed by Franco et al. (Reference Franco, Rubini and Vecchi1982), but the presence of F D can be deduced from the occurrence of at least one homozygous M male in all autosomal populations and the occurrence of XY females and YY males in mixed populations (Franco et al., Reference Franco, Rubini and Vecchi1982), implying that 25 years ago F D (or a similar genetic element) was present across the entire range of Italy, as it is now. However, we did find F D in Switzerland, where it was not detected before 1982, suggesting that the F D factor has spread northwards slightly.
4. Discussion
Our results show that autosomal M factors have not spread northwards in Europe over the last 25 years, in contrast to what was predicted by Franco et al. (Reference Franco, Rubini and Vecchi1982). One may argue that we have overlooked low frequencies of autosomal M factors in Switzerland and Germany due to insufficient sample size. Although this may be true, very low frequencies of autosomal factors still support the hypothesis that the standard XY system is not being replaced by autosomal factors in northern populations. In line with our results, we suggest that after their initial spread in southern localities (see Franco et al., Reference Franco, Rubini and Vecchi1982), autosomal M factors reached a stable distribution.
Our results indicate that some factors prevent the spread of autosomal M in populations north of Italy. In the transect we studied, the Alps may be considered as a barrier, although the biology of the housefly and its ease of spread with human transportation seem to preclude this physical barrier as being important for the potential long-term spread of autosomal M factors. In fact, the presence of the F D factor north of the Alps and the M factor on autosome II in flies collected in eastern France in 2004 (results not shown) suggests that geographical barriers do not prevent the northward spread of autosomal M factors. More likely, some climatic factors are responsible for the stability of the distribution of M. The most obvious climatic factor related to latitude is temperature, which has been shown to be a strong predictor of the frequencies of different sex-determining factors in the housefly worldwide (Feldmeyer et al., submitted). However, it is not obvious how temperature might influence the evolution and distribution of SD mechanisms (discussed in detail in Feldmeyer et al., submitted).
Our statistical analysis reveals an effect of temperature, but also a significant interaction between temperature and latitude on the frequency of autosomal SD factors (Table 4). The interaction stems from the fact that at higher latitudes temperature has a positive effect on the frequencies of autosomal SD factors, whereas the opposite pattern is present at lower latitudes (not shown). This may suggest that autosomal SD factors reach the highest frequencies at intermediate temperatures. However, autosomal SD factors have been found at high frequencies in places where average temperatures are higher than at our sampling sites (Feldmeyer et al., submitted). A more likely explanation is that temperature interacts with other climatic factors (such as humidity) that could be correlated with latitude (and altitude) in our study area. This could also explain why an M factor on autosome III and F D have been found at locations in England where the yearly range of temperatures is similar to that in Germany and Switzerland (Denholm et al., Reference Denholm, Franco, Rubini and Vecchi1985; data on temperatures from WORLDCLIM, not shown). Additionally, M factors located on different autosomes may be differently affected by temperature.
It has also been proposed that autosomal M factors have spread due to their linkage with insecticide resistance genes (Kerr, Reference Kerr1970; Franco et al., Reference Franco, Rubini and Vecchi1982), since the isolation of autosomal M factors coincided with the appearance of insecticide resistance in natural populations of the housefly (Tomita & Wada, Reference Tomita and Wada1989b). Also, in a number of resistant populations autosomal M males have been found (Tsukamoto, Reference Tsukamoto, Georghiou and Saito1983) and one laboratory experiment showed replacement of standard XY males by autosomal M males after several generations of selection for DDT resistance (Kerr, Reference Kerr1970). However, even though linkage with insecticide resistance genes could facilitate spread of autosomal M factors, it is not clear how it could contribute to the clinal distribution of SD factors in the housefly. One could argue that in warmer climates more generations of flies are produced and more applications of insecticides are used, allowing faster spread of M factors linked with insecticide-resistant genes. However, since pesticides have been used throughout Europe for decades and resistance genes are widespread also in northern populations (Keiding, Reference Keiding, Watson and Brown1977, Reference Keiding1999), one would expect that M factors would be increasing in frequency, although more slowly, in the north also. As we showed in this study, this is not the case. Another argument is that there is no correlation between the frequency of autosomal M males and insecticide resistance in housefly populations from the eastern United States (Hamm et al., Reference Hamm, Shono and Scott2005). Therefore, linkage with insecticide resistance genes might explain the spread of autosomal M factors in some cases, but it seems unlikely to provide a general explanation for the clinal distribution of SD factors in the housefly.
Interestingly, autosomal M factors are not fixed in most populations and multiple factors on several or even all chromosomes can be maintained in a single population. This polymorphism was one of the reasons underlying the opinion of earlier researchers that the sex-determining mechanism in the housefly is in a transient state (e.g. Franco et al., Reference Franco, Rubini and Vecchi1982; Denholm et al., Reference Denholm, Franco, Rubini and Vecchi1985; Tomita & Wada, Reference Tomita and Wada1989b). However, theoretical models reveal that such a polymorphism can be stable not only for specific fitness values of different genotypes (Bull & Charnov, Reference Bull and Charnov1977; Jayakar, Reference Jayakar1987), but also when different genotypes have the same viability and fertility (Kozielska et al., Reference Kozielska, Pen, Beukeboom and Weissing2006). Therefore, the conditions for a stable polymorphism may be much less restrictive than previously thought, and it may well be that the multifactorial SD system of the housefly is stable.
Unfortunately, we do not have data on the frequencies of different autosomal M factors in the past to see whether these frequencies have changed. Franco and colleagues (Reference Franco, Rubini and Vecchi1982) did not find any M factors located on autosomes I, IV or V, but they do not provide the number of males investigated. If these factors were present in the past at low frequencies as they are now (Table 3), Franco et al. (Reference Franco, Rubini and Vecchi1982) might not have detected them in small sample sizes. They reported that M was more common on autosome III than on autosome II. The same pattern is seen in this study and several others (Tomita & Wada, Reference Tomita and Wada1989b; Denholm et al., Reference Denholm, Rubini, Rovati and Vecchi1990; Hamm et al., Reference Hamm, Shono and Scott2005; except for Tanzanian populations, Feldmeyer et al., submitted.). This suggests that M on autosome III confers the largest fitness gain to its bearer, but this may only be a conditional effect (e.g. frequency- or temperature-dependent) since the M on autosome III did not replace other M factors in recent decades in the Italian populations.
Another explanation for the high polymorphism in genomic location of M factors is that the M factor is part of a transposable element, as is known for the M factor in Megaselia scalaris (Traut & Willhoeft, Reference Traut and Willhoeft1990). In this species transposition rate differs depending on the chromosome on which M is located (Green, Reference Green1980). This might not only explain why M factors are more common on some autosomes than others, but also the clinal distribution of M factors, since transposition rate is known to be dependent on temperature and often increases with increasing temperature (Lampe et al., Reference Lampe, Grant and Robertson1998; Ohtsubo et al., Reference Ohtsubo, Genka, Komatsu, Nagata and Tsuda2005; but see Hashida et al., Reference Hashida, Kitamura, Mikami and Kishima2003). Molecular studies are necessary to establish whether the M factor is always the same gene located on a transposable element or whether M factors on different chromosomes are different genes blocking the female-determining factor F (see Dübendorfer et al., Reference Dübendorfer, Hediger, Burghardt and Bopp2002).
Our crosses suggest that the frequency of the Y chromosome has increased over recent decades in some Italian populations. We found an M factor on the sex chromosomes in some populations that were described as purely autosomal by Franco and colleagues (Reference Franco, Rubini and Vecchi1982; Fig. 3). It is difficult to assess what the cause of these changes in particular populations is; some local factors may be involved. For population IT5, the difference between past and present frequencies of XX males might reflect the fact that we could not locate accurately the sampling site of Franco and colleagues (Reference Franco, Rubini and Vecchi1982; see Section 2). Moreover, it should be noted that due to the absence of visible markers on the sex chromosomes of the housefly, our crosses did not allow us to determine whether the M factor was present on the Y or on the X chromosome (as found in England: Denholm et al., Reference Denholm, Franco, Rubini and Vecchi1983, Reference Denholm, Franco, Rubini and Vecchi1985). Without additional information, the data obtained from the crosses could easily lead to the incorrect classification of XXM males as XY males. Therefore, we performed additional cytological investigations, using orcein staining, a standard technique employed in cytological studies of the housefly (Hiroyoshi, Reference Hiroyoshi1964; Franco et al., Reference Franco, Rubini and Vecchi1982; Denholm et al., Reference Denholm, Franco, Rubini and Vecchi1983, Reference Denholm, Franco, Rubini and Vecchi1985). Our preliminary results (not shown) confirm that males from the northernmost populations (GE1, GE2, SW and IT3) are of karyotype XY. Unfortunately, we could not unambiguously distinguish between XX, XY and YY karyotypes in the other populations, because the length polymorphism of the housefly sex chromosomes (also know from other strains: Boyes et al., Reference Boyes, Paterson and Corey1964; Boyes, Reference Boyes, Wright and Pal1967; Milani, Reference Milani1971; Franco et al., Reference Franco, Rubini and Vecchi1982; Denholm et al., Reference Denholm, Franco, Rubini and Vecchi1983, Reference Denholm, Franco, Rubini and Vecchi1985; Hediger et al., Reference Hediger, Niessen, Müller-Navia, Nöthiger and Dübendorfer1998) did not allow a reliable distinction between X and Y chromosomes. Therefore, we cannot exclude the possibility that the X chromosome (rather than the Y chromosome) bears the M factor in the southern populations.
In conclusion, even if the distribution of the Y chromosome in European populations is difficult to assess, our main conclusion that autosomal M factors have not spread northwards in the last 25 years still holds. This suggests that the polymorphism of SD factors in natural housefly populations is not transient but stable. Additional studies, at both the ecological and the molecular level, are required to unravel the factors responsible for the stable coexistence of various SD factors. Undoubtedly, better understanding of the housefly SD system will also provide general insights into the evolution of sex determination, which is still poorly understood in other taxa as well.
We are grateful to a number of people who contributed to the project: Bernd Grillenberger for help during the fieldwork and Sezin Daðdeviren with molecular work; Laas Pijnacker for the cytological work; Monika Hediger and Daniel Bopp (Developmental Biology, University of Zurich) for providing the marker strains and molecular primers. Financial support for the fieldwork was received from the Schure-Beijerinck-Popping Funds of the Royal Netherlands Academy of Arts and Science (KNAW) in the form of a Field Grant to M.K.










