1. Introduction
Nasonia vitripennis is a widely used parasitoid wasp model system to study major topics such as genetics, development, ecology, and behavior (Beukeboom & Desplan, Reference Beukeboom and Desplan2003; Lynch, Reference Lynch2015). The first sequencing of its genome has been published in 2010 (Werren et al., Reference Werren, Richards, Desjardins, Niehuis, Gadau, Colbourne, Beukeboom, Desplan, Elsik, Grimmelikhuijzen, Kitts, Lynch, Murphy, Oliveira, Smith, Van Zande, Worley, Zdobnov, Aerts and Gibbs2010). Recently the development of new technologies has pushed the model forward. For example, knock out of genes by the CRISPR/Cas9 technology has been reported but has at the moment not been widely used (Li et al., Reference Li, Au, Douglah, Chong, White, Ferree and Akbari2017) due to low survivability of embryos. Therefore, knockdown of genes by RNA interference (RNAi) or by parental RNAi (pRNAi) is still the most popular method to study gene function and maternal transcript provision in N. vitripennis (Dalla Benetta et al., Reference Dalla Benetta, Antoshechkin, Yang, Nguyen, Ferree and Akbari2020; Lynch & Desplan, Reference Lynch and Desplan2006; Wang et al., Reference Wang, Rensink, Fricke, Riddle, Trent, van de Zande and Verhulst2020; Werren et al., Reference Werren, Loehlin and Giebel2009). However, in other Hymenoptera such as the honeybee, RNAi has been reported to have undesirable off-target effects, for example when using GFP dsRNA as a control (Jarosch & Moritz, Reference Jarosch and Moritz2011; Nunes et al., Reference Nunes, Aleixo, Barchuk, Bomtorin, Grozinger and Simoes2013). Here we investigate the potential for off-target effects of using GFP double-strand RNA (dsRNA) as non-target control in N. vitripennis RNAi experiments.
2. Objective
As RNAi has become a widely used technique in N. vitripennis (Lynch, Reference Lynch2015), it is of importance to identify potential pitfalls generated by the technique. Also, with the now affordable cost of RNA-sequencing technologies, RNAi experiments can be coupled with genome-wide transcriptomic analyses. Therefore, identifying the potential off-target effects of RNAi in N. vitripennis will help researchers to design appropriate controls for their experiments and avoid confusing results or following false positive leads. To this aim, we tested the off-target effect of a non-specific RNAi response by comparing the effect of injecting dsRNA against the non-target GFP together with water-injected or uninjected controls at whole transcriptome level.
3. Methods
Second instar male larvae were injected with either GFP dsRNA (GFP-i), water, or were not injected but otherwise treated the same. Each condition was analyzed in triplicate using five males per sample. The treated males were collected approximately five days later at the white pupal stage and RNA was extracted with Quick-RNA Tissue/Insect Kit according to manufacturer’s protocol (ZymoResearch – R2030). Libraries were prepared with a custom protocol and 150 bp paired-end sequencing was achieved on a HiSeqX (Illumina) by Novogene (Novogene, HK company limited). From the fastQ files, read counts per gene were retrieved using GeneCounts quantification method from STAR (Dobin et al., Reference Dobin, Davis, Schlesinger, Drenkow, Zaleski, Jha, Batut, Chaisson and Gingeras2013) version 2.6.1b and the Nvit_psr_1.1 N. vitripennis genome version with RefSeq annotation GCF_009193385.2 as reference. Differential expression analysis was calculated with DESeq2 (Love et al., Reference Love, Huber and Anders2014) version 1.20.0. Gene Ontology analyses were performed using DAVID bioinformatics resources (Huang da et al., Reference Huang da, Sherman and Lempicki2009) version 6.8. For comparison with the Nunes et al. microarray data from honeybee, only genes found affected in at least two experiments were used (Nunes et al., Reference Nunes, Aleixo, Barchuk, Bomtorin, Grozinger and Simoes2013).
Complete detailed description of the methods is available as supplemental material.
4. Results
Full statistics of bioinformatics analysis are presented in Table S1 and show consistent high quality data in all samples. Pearson correlation analysis shows that the samples have a high correlation coefficient and cluster in three different groups but not based on experimental conditions (Figure 1A). Principal component analysis also revealed no apparent clustering of the different samples based on experimental conditions, except for one replicate of uninjected control segregating away on the PC2 (Figure 1B). Altogether, these results suggest few differences between samples.
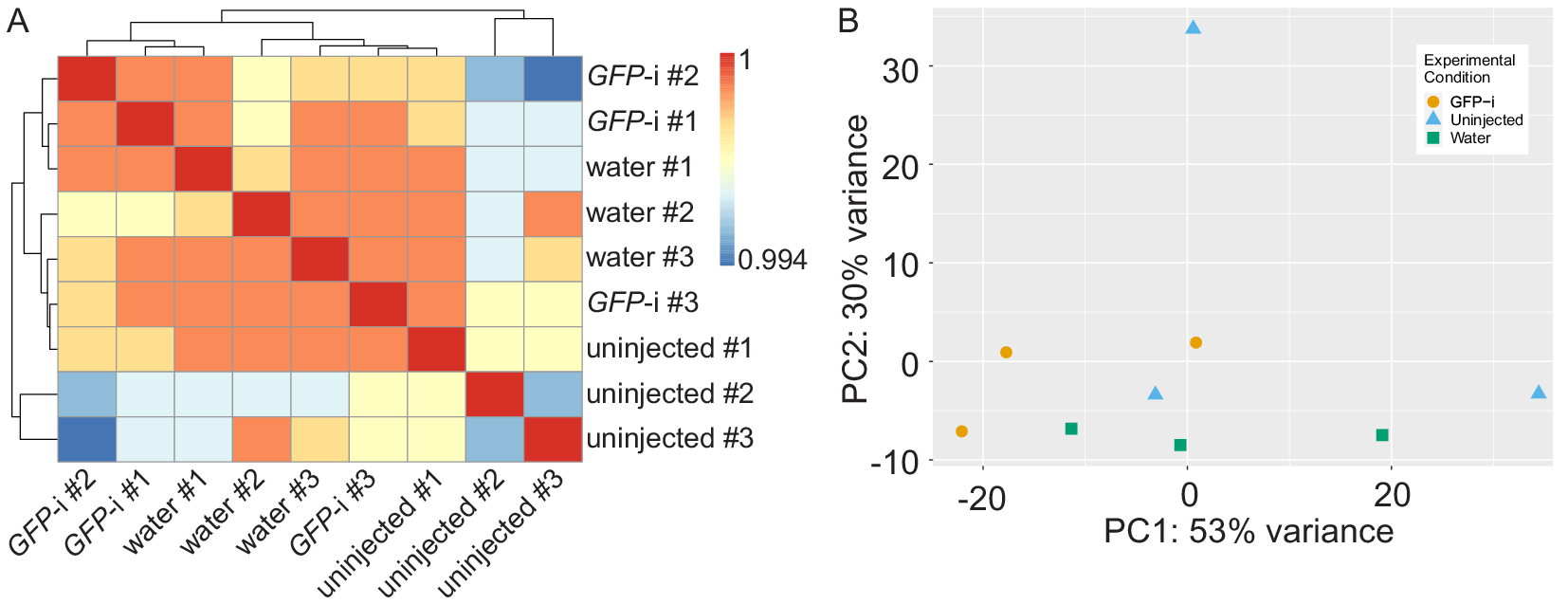
Figure 1. Analysis of RNA-sequencing sample reproducibility. (A) Heatmap of Pearson correlation coefficient (r) of GFP-i, water-injected, and uninjected samples. (B) Principal component analysis of GFP-i (orange dots), uninjected (blue triangles), and water-injected (green squares) samples.
Differential expression analysis between uninjected and water-injected samples with an adjusted P-value threshold of (P-adj) < 0.05, revealed that only 46 genes are differentially expressed (Table S2). Therefore, we decided to pool these two conditions into one control group. Differential expression analysis between GFP-i and control samples with P-adj < 0.05 revealed 518 differentially expressed genes (DEG), of which 71 have a higher expression and 447 a lower expression (Figure 2 and Table S3). Gene Ontology analysis on these 518 DEGs identified enrichment for “microtubule-based process”, “cytochrome-c oxidase activity” and “oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor”.
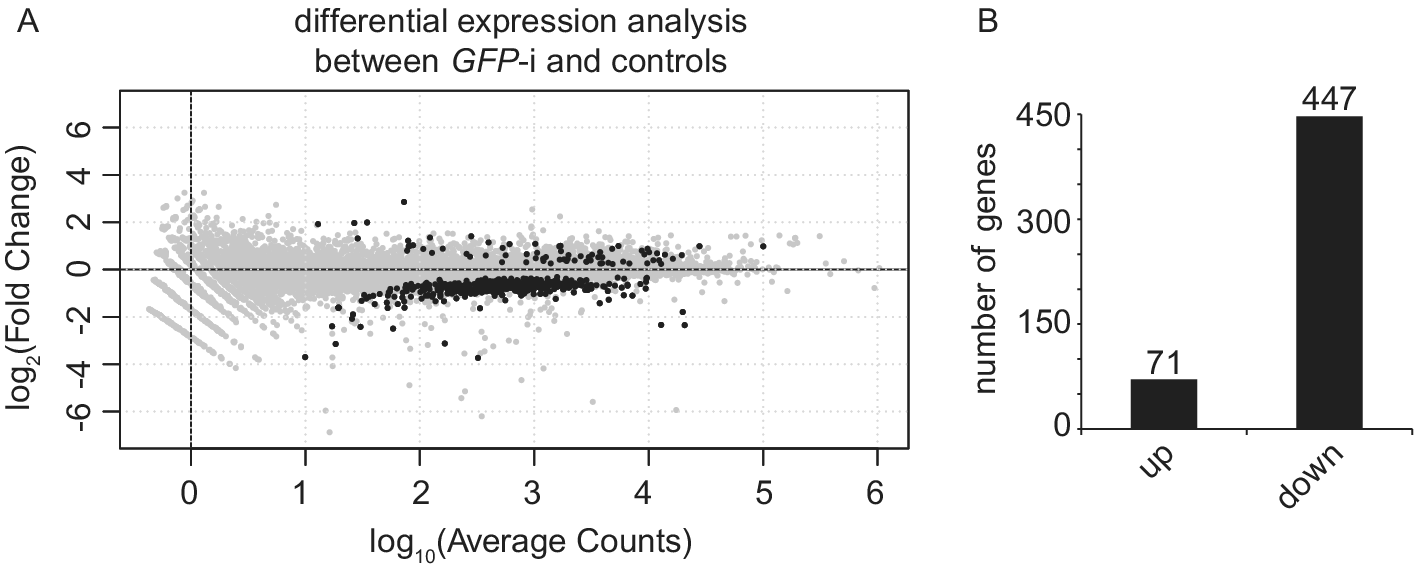
Figure 2. Injection of GFP dsRNA induces changes in gene expression when compared with controls. (A) MA plot showing the fold change (log2-transformed) between gene expression in GFP-i and control (uninjected and water-injected combined) samples as a function of the normalized average count between the two conditions (log10-transformed), as calculated with DEseq2. Genes with significantly differential expression (P-adj < 0.05) are showed in black. For GFP-i and control samples, three and six biological replicates were used, respectively. (B) Bar plot quantifying genes found more expressed (log2FC > 0 and P-adj < 0.05) or less expressed (log2FC < 0 and P-adj < 0.05) after differential expression analysis.
To find regions in the N. vitripennis genome with a potential GFP sequence similarity causing the off-target effects, we aligned the GFP sequence used in our experiment against the N. vitripennis genome. Blastn identified 17 hits with an alignment length ranging from 18 to 46 nucleotides and between 78% to 100% identity (Table S4). Aside from LOC100122071, none of these genes aligned gapless for more than 17 nucleotides. Of these 17 genes, only LOC103318004 is differentially expressed in our RNAseq experiments, with a reduced expression in both water-injected and GFP-i samples (log2FC = −0.76 with P-adj = 0.013 and log2FC = −1.03 with P-adj = 0.006 respectively). We also used the Nasonia specific RNAi off-target prediction tool (Davies & Tauber, Reference Davies and Tauber2015) and it did not find any matching 19-mers of GFP sequence.
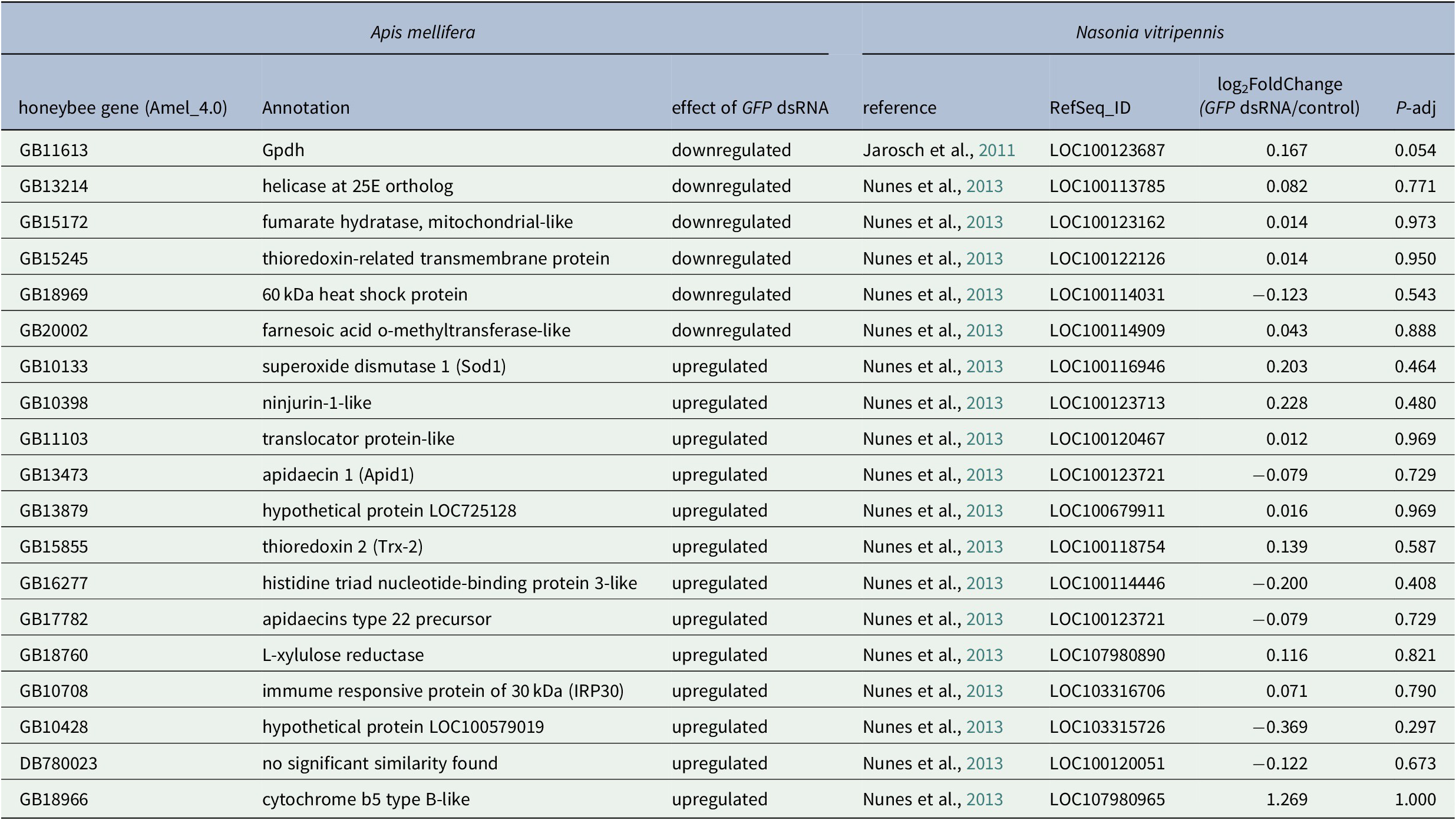
Finally, we compared our results with microarray data obtained in another Hymenoptera, the honeybee Apis mellifera. Nunes et al. identified 1,416 differentially expressed genes after GFP dsRNA injection across three different experimental conditions, with high variations between samples and only 18 genes differentially expressed in at least two conditions (Nunes et al., Reference Nunes, Aleixo, Barchuk, Bomtorin, Grozinger and Simoes2013). None of these 18 genes were in our N. vitripennis DEG list (Table 1).
Table 1. Comparison between the effect of GFP dsRNA injection in Apis mellifera and Nasonia vitripennis shows that none of the DE genes found in the two A. mellifera studies are differentially expressed in N. vitripennis.
5. Discussions
Comparison of uninjected and water-injected samples did not reveal a major response caused by the injection procedure itself five days before sampling. However, among the 46 DEGs, LOC100117489 (pyrazinamidase/nicotinamidase) and LOC100120201 (vanin-like protein 2) have been previously linked to oxidative stress in Drosophila and human (Balan et al., Reference Balan, Miller, Kaplun, Balan, Chong, Li, Kaplun, VanBerkum, Arking, Freeman, Maiese and Tzivion2008; Bartucci et al., Reference Bartucci, Salvati, Olinga and Boersma2019) and may suggest some level of stress response upon injection.
Our RNAseq experiments revealed that 518 out of 15,430 total genes (3.4%) are differentially expressed in whole pupae in response to GFP-i with P-adj < 0.05. Among the 447 lower expressed genes, 33 are coding for microtubules or microtubule-associated proteins, predicted to control sperm functions. Also, four cytochrome C oxydase subunits show a decrease in expression. In addition to be possible markers of apoptotic stress, cytochrome C oxydases have been linked to sperm differentiation in Drosophila (Arama et al., Reference Arama, Agapite and Steller2003). Finally, LOC103318004, which is predicted to encode a kelch-like protein 10, showed some sequence similarity with the GFP sequence used in this study. However these two sequences aligned on less than 19 nucleotides without gap, which was described as the minimum consensus length for RNAi (Elbashir et al., Reference Elbashir, Lendeckel and Tuschl2001). LOC103318004 homologs have been linked to gamete development in both Drosophila and mice (Hudson & Cooley, Reference Hudson and Cooley2010; Yan et al., Reference Yan, Ma, Burns and Matzuk2004). LOC103318004 is less expressed in GFP-i samples compared to controls, but, surprisingly, also in water-injected samples when compared to uninjected controls. Therefore, it is unclear whether the decrease in expression of this gene is due to injection stress or is a sequence-specific GFP-i effect, or both. As dsRNA was injected in the abdomen, activation of the RNAi response in this region could affect spermatogenesis. Similar observations have been made in female N. vitripennis, which show some reduced fertility after water or dsRNA injection at pupal stage (Geuverink et al., Reference Geuverink, Rensink, Rondeel, Beukeboom, van de Zande and Verhulst2017).
6. Conclusions
Our transcriptomic comparison of GFP non-specific RNAi with water and uninjected controls revealed moderate off-target effects in male N. vitripennis, potentially affecting spermatogenesis. Only in one case we were able to relate this effect to endogenous GFP sequence similarity in N. vitripennis, and this leads us to assume that GFP-i does not cause specific off-target effects, but instead results in a general response to dsRNA. As we injected into the abdomen, close to the reproductive organs, it could additionally lead to reduced fertility in males. It should be noted that our study focuses on only one sex at one developmental time and results may differ under different experimental conditions.
The lack of overlap between DEGs after RNAi activation in Nasonia and honeybees suggests that off-target effects cannot be generalized across Hymenoptera. The specific effects of RNAi activation and off-target effects should be studied on a species-specific basis. According to our results, GFP seems a suitable non-targeting negative control for RNAi experiments in N. vitripennis as it appears to elicit a general non-specific response against dsRNA, and sets a baseline reference for the target-specific knockdown. However, any RNAi study involving the genes we show to be affected in this study should exercise caution in drawing conclusions and may be safer by using a different non-target control.
Author Contributions
Y.W., J.R. and E.C.V. designed the study. Y.W. performed experiments. J.R. analyzed data. J.R. wrote the initial manuscript draft, all authors revised the draft and approved the final version. E.C.V. acquired funding.
Funding Information
Part of the work was funded by the Innovative Research Scheme of the Dutch Research Council (NWO) granted to ECV (016.Vidi.189.099).
Publishing Ethics
The authors confirm that
1. the manuscript has been submitted only to the journal – it is not under consideration, accepted for publication or in press elsewhere. Manuscripts may be deposited on pre-print servers;
2. all listed authors know of and agree to the manuscript being submitted to the journal;
3. the manuscript contains nothing that is abusive, defamatory, fraudulent, illegal, libelous, or obscene.
Conflict of Interest
The authors declare no conflict of interest.
Data Availability
The sequencing data have been submitted to GEO under accession number GSE153268.
Supplementary Materials
To view supplementary material for this article, please visit http://dx.doi.org/10.1017/exp.2020.67.





Comments
Comments to the Author: Using RNA-Seq, the authors analyzed the potential undesired effects of dsRNA-GFP as a control in RNAi experiments on the Nasonia vitripennis wasp species. Compared to the previous version of the manuscript, there are many improvements, especially with the use of a less stringent p-value that made it possible to perceive a greater number of genes impacted by GFP-i. Overall, I appreciate the current version of the manuscript and I just have a few questions/ suggestions about it.
In the files I received for analysis there are two titles, but I believe that the correct and most appropriate one is: Effect of using Green Fluorescent Protein double-stranded RNA as non-target negative control in Nasonia vitripennis RNA interference assays.
Based on the GFP-primers sequences, the reported dsRNA size (460 bp) and the Emerald GFP CDS sequence, I believe the target fragment used for dsRNA-GFP synthesis is the one below and should be included in the supplementary material:
5 ‘ - GTGACCACCTTGACCTACGGCGTGCAGTGCTTCGCCCGCTACCCCGACCACATGAAGCAGCACGACTTCTTCAAGTCCGCCATGCCCGAAGGCTACGTCCAGGAGCGCACCATCTTCTTCAAGGACGACGGCAACTACAAGACCCGCGCCGAGGTGAAGTTCGAGGGCGACACCCTGGTGAACCGCATCGAGCTGAAGGGCATCGACTTCAAGGAGGACGGCAACATCCTGGGGCACAAGCTGGAGTACAACTACAACAGCCACAAGGTCTATATCACCGCCGACAAGCAGAAGAACGGCATCAAGGTGAACTTCAAGACCCGCCACAACATCGAGGACGGCAGCGTGCAGCTCGCCGACCACTACCAGCAGAACACCCCCATCGGCGACGGCCCCGTGCTGCTGCCCGACAACCACTACCTGAGCACCCAGTCCGCCCTGAGCAAAGACCCCAACGAGA - 3’
Lines 28-29: replace “for example when using GFP coding sequence as a control” by “for example when using GFP dsRNA as a control”.
The authors did not report in the main text or supplementary material how many micrograms were used to prepare the RNA-Seq libraries (for the enrichment of mRNAs and first strand cDNA synthesis).