INTRODUCTION
Extraintestinal infections with multidrug-resistant (MDR) E. coli are increasingly recognized in dogs and can result from the endogenous transfer of bacteria from the canine patient's faecal microbiota [Reference Ball1, Reference Donskey2]. The gastrointestinal tract is an important reservoir for MDR Gram-negative bacilli and intestinal colonization often precedes clinical infection [Reference Cookson3], contamination of the environment [Reference Johnson, Stell and Delavari4] and subsequent colonization of other animals or humans [Reference Damborg, Nielsen and Guardabassi5].
The gastrointestinal tract is also an important site for horizontal transfer of resistance genes between pathogens and commensal organisms, which can be exacerbated by selective pressure exerted by antimicrobial use [Reference Donskey2, Reference Summers6]. In humans, the prevalence of rectal MDR E. coli appears to be increased by antimicrobial drug use [Reference Filius7–Reference Harris9] as well as factors such as hospitalization [Reference Filius7], time in intensive care units (ICUs) [Reference Filius7], disease conditions [Reference Harris9], and age [Reference Harris9]. In contrast, determinants of E. coli colonization of the gastrointestinal tract in hospitalized dogs have received limited attention to date [Reference Murphy10, Reference Ogeer-Gyles11].
MDR E. coli were involved in an outbreak of extraintestinal infections in dogs at The University of Queensland Small Animal and Veterinary Teaching Hospital (UQVTH) from 2000 to 2002 [Reference Sidjabat12]. In a case-series of 37 dogs with MDR E. coli and Enterobacter extraintestinal infections, predisposing disease condition, prior antimicrobial use, duration of hospitalization, and type of surgical procedure were identified as potential risk factors [Reference Gibson13]. Characterization of initial isolates from an infection control programme to limit MDR E. coli transfer between animals revealed the same two distinct clonal groups of MDR E. coli in rectal swabs from hospitalized dogs that had previously been identified in extraintestinal infections [Reference Sidjabat14], with clonal group 1 (CG1) corresponding to E. coli phylogenetic group A and clonal group 2 (CG2) corresponding to phylogenetic group D [Reference Sidjabat15].
Identification and isolation of dogs that are at a greater risk of being colonized with MDR bacteria could be an important mitigation strategy to prevent the introduction of MDR pathogens into the veterinary hospital environment. In the only previous study on risk factors for carriage of MDR E. coli in a veterinary setting, the study population was restricted to hospitalized dogs in ICUs and only two risk factors were assessed: duration of hospitalization and antimicrobial agents administered [Reference Ogeer-Gyles11]. The risk of positive rectal colonization with MDR E. coli in canine patients increased with time hospitalized in ICU, and prior fluoroquinolone use was identified as a risk factor for isolation of quinolone-resistant E. coli [Reference Ogeer-Gyles11].
In the current study, potential risk factors for rectal colonization with MDR E. coli in dogs on admission to a large veterinary teaching hospital including prior treatments, hospitalizations and interventions were assessed using the data collected at admission during the entire infection control programme at the UQVTH. In addition, the characteristics of the MDR E. coli isolates obtained during this infection control programme are described.
METHODS
Study overview
This study was a retrospective prevalence-based case-control study using information collected during an infection control programme at the UQVTH, a first opinion and referral hospital in Brisbane, Australia, between 7 August 2000 and 15 November 2002. As part of the programme, where possible, rectal swabs were taken from dogs on admission to hospital. The unit of interest for this study was the individual admission, and cases and controls were selected at the admission level. Frequencies of exposure to potential risk factors were compared between admissions that were rectally colonized with MDR E. coli and admissions that were not colonized.
Case and control selection
Admissions were selected from the dates between 1 March 2001 and 30 October 2002. This date range was chosen as, during this phase of the infection control programme, compliance with rectal swabbing was highest. Compliance was assessed for one week of each month. During the 21 weeks from this period that were assessed, there were 396 admissions, of which 285 (72%) were swabbed for MDR E. coli on either day of admission and/or the following day, in contrast to the 6 weeks prior to and after this period, where only 5/153 (9·4%) and 6/19 (31·6%) of admissions were swabbed, respectively. Dogs were considered to be admitted to hospital if they were moved from a consultation room and caged in the hospital for at least one night. Rectal swabs were cultured on MacConkey agar containing enoxacin (5 mg/ml) and gentamicin (5 mg/ml) (MCAEG) as previously described [Reference Sidjabat14]. Isolates were stored in Luria–Bertani broth with 15% (v/v) glycerol.
Admissions were selected from dogs swabbed on either the day of admission and/or the following day. Case admissions were selected from those where swabs were positive (growth on MCAEG) on initial admission samples. Control admissions were selected from dogs that had a negative rectal swab for one or both dates sampled. For dogs from which MDR E. coli was isolated at more than one admission during the study period, only the first admission was enrolled. Dogs were eligible for selection as controls at more than one admission and were also eligible for selection as controls at admissions before MDR E. coli was isolated during the study period, but were ineligible for selection as controls at admissions after MDR E. coli was isolated. For each case admission, one control admission was randomly selected using computer-generated random numbers from client-owned dogs that had a negative rectal swab collected on the same admission date. If there was no admission eligible for selection as a control on the same date as the case admission, a control was randomly selected from an adjoining day. If there was no eligible control within a week either side of the case, the case was excluded. Both primary-care and referral admissions were eligible for selection. Only client-owned dogs were eligible for selection.
Throughout this time, 116 admissions were eligible for enrolment as cases. Of these, nine admissions were excluded due to incomplete or incorrect rectal swab data. Admissions were then excluded if the dog's clinic file was missing (n=24) or no appropriate time-matched control was available (n=1). Fourteen selected control admissions also had to be replaced as there was no clinic file available. This resulted in 82 case admissions and 82 time-matched control admissions (matched on admission date) from 157 dogs. Six dogs provided more than one study admission: one dog provided two control admissions; four dogs provided a control admission then case admissions at a later admission; and one provided two control admissions then a case admission.
Potential risk factor data collection
Exposure to potential risk factors during the 42 days preceding the study admission date (defined as the pre-admission period), were assessed. This time period was selected based on durations of rectal colonization in dogs following experimental infection with MDR E. coli [Reference Trott16]. In this study, dogs which had prior antimicrobial therapy remained colonized with MDR E. coli for a mean of 26 days, with the longest colonization period being 38 days. Potential risk factors were selected based on veterinary adaptations of related human literature. Data for each risk factor of interest was collected by examining hospital records as well as the records from referring veterinary practices. Potential risk factors were categorized as shown in Table 1.
Table 1. Potential risk factors for rectal colonization with MDR E. coli in dogs on admission to hospital that were assessed in a retrospective, prevalence-based case-control study; exposures to time-varying factors were for the 42-day period prior to admission (the ‘pre-admission period’)
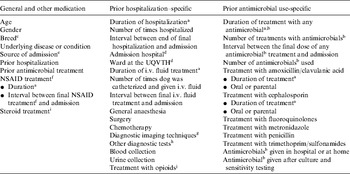
a Cumulative days hospitalized or treated.
b Antimicrobials included amoxicillin/clavulanic acid, ampicillin, first generation cephalosporin, fluoroquinolones (enrofloxacin, orbifloxacin), aminoglycoside (gentamicin), carbapenem (imipenem), lincosamide (clindamycin), metronidazole, tetracycline (doxycycline) and trimethoprim/sulfonamide.
c Mixed breed dogs were placed into a purebred grouping for analysis if a main breed was listed. Breed was analysed using two breed categorizing methods. A genetic breed category based on the genetic structure of the purebred domestic dog [Reference Parker27] and breed grouped according to the Australian National Kennel Council (ANKC) database [28].
d Study dogs were classified as having been admitted to The University of Queensland Veterinary Teaching Hospital (UQVTH), other veterinary hospitals in the surrounding area or a combination of the UQVTH and another veterinary hospital.
e Source, if records showed the dog had been referred from another veterinary hospital it was referred, otherwise it was primary care.
f Non-steroidal anti-inflammatory drugs (NSAIDs) included carprofen, meloxicam and piroxicam.
g Diagnostic imaging techniques included radiography, echocardiography, ultrasonography and computer tomography.
h Other tests included aspirates (chest, joints and wounds), CSF tap, endoscopy, ear swabs and tracheal washes.
i Steroidal anti-inflammatories included dexamethasone and prednisolone.
j Opioids included morphine, fentanyl and buprenorphine.
Data analyses
Associations between potential risk factors and rectal colonization with MDR E. coli were assessed using logistic regression. Maximum-likelihood logistic regression models were fitted using Stata, version 10.1 (StataCorp, USA). As six dogs each provided more than one admission, a random effect of dog was fitted in all maximum-likelihood logistic models. As controls were effectively frequency-matched rather than 1:1 matched, and so that we could allow for a random effect of dog, unconditional models were used. Overall significance of variables was assessed using likelihood ratio P values and significance of individual levels of risk factors (relative to the reference level) was assessed using Wald P values. Variables with no control admissions for one level of the potential risk factor were assessed using exact logistic regression models, fitted using LogXact 8 (Cytel Inc., USA). P values for estimates for each level of these risk factors (relative to the reference level) were calculated as two times the one-sided exact P values. Exact probability P values were used for hypothesis testing of the overall significance of these risk factors. Odds ratios (OR) for these exposure variables were obtained using the median unbiased estimator [Reference Hosmer and Lemeshow17].
Potential risk factors were analysed using univariable analysis before and after adjustment for admission month. For all except two antimicrobial-specific factors (interval between the final dose of any antimicrobial and admission and duration of prior amoxicillin/clavulanic acid treatment), the estimated OR changed by <30% after adjustment for admission month. Accordingly, admission month was not fitted in multivariable models and univariable results were used. These variables were subsequently further explored by fitting with the final maximum-likelihood model (other than prior antimicrobial treatment for the former) and admission month.
This study took place at the same time as an infection control programme. This may have altered the risk of prior hospitalization at the UQVTH for admissions later in the study period. Therefore, the risk of prior hospitalization was also analysed by fitting this variable with prior hospitalization at the UQVTH separated into hospitalization during the first half (1 March 2001 to 30 November 2001) and second half (1 December 2001 to 18 October 2002) of the study.
After univariable analysis, all variables except those requiring analysis using an exact model that had overall likelihood ratio P values <0·2 were assessed using multivariable modelling. Each of the variables that were not specific to hospitalization or antimicrobials (detailed in Supplementary Table 1, available online) was fitted using a forward selection approach with variables sequentially fitted in ascending order based on P value. Variables with overall P values were sequentially excluded before further variables were fitted. Once excluded, variables were not eligible for re-inclusion.
An additional model was then developed using the subset of dogs that had previously been hospitalized to investigate hospitalization-specific variables (detailed in Supplementary Table 1) using the same forward selection process with prior antimicrobial treatment (the only variable retained in the previous model) forced into all models.
To further investigate effects of antimicrobial use, antimicrobial-specific variables (detailed in Supplementary Table 1) were then assessed using the same forward selection process with prior diagnostic imaging techniques and prior duration of hospitalization (the only variables retained in the previous model) forced into all models. Because no control dogs had received fluoroquinolones and because fluoroquinolones were only given to dogs which were hospitalized in the pre-admission period, prior fluoroquinolone use was further examined using exact multivariable analysis after adjusting for duration of hospitalization and diagnostic imaging techniques and prior use of non-fluoroquinolone antimicrobials, using only dogs that had been hospitalized.
The fit of the final maximum-likelihood logistic model (prior antimicrobial treatment, prior diagnostic imaging techniques and prior duration of hospitalization) was assessed without the random effect of dog using the Hosmer–Lemeshow goodness-of-fit test and by comparing observed to expected numbers of cases and controls for five groups based on predicted probabilities. The discriminatory ability of the final maximum-likelihood model without the random effect of dog was assessed using the area under the receiver operator characteristics (ROC) curve and by assessing sensitivity and specificity of the model at varying probability cut points.
Characterization of MDR isolates
One isolate was characterized from 58/82 case dogs. Isolates had not been stored for 24 case dogs: these were mostly obtained from cases presenting towards the end of the infection control study. Disc diffusion susceptibility testing for amoxicillin/clavulanic acid, cefotaxime, cefoxitin, chloramphenicol, enrofloxacin and spectinomycin was performed on isolates using methods described in CLSI guidelines [18]. Isolates were also confirmed to be AmpC β-lactamase-producing E. coli and divided into clonal groups based on results of a multiplex PCR for E. coli uspA, bla CMY and a class 1 integron-associated dfra17-aadA5 [Reference Sidjabat14]: MDR E. coli CG1 isolates are positive for all three genes and CG2 isolates positive for uspA and bla CMY only.
RESULTS
Univariable analyses
Male entire dogs were at greater risk of colonization than were other gender classes [OR 4·94, 95% confidence interval (CI) 1·58–15·39, P=0·006]. Breed was not significantly associated with presence of MDR E. coli under either of two breed categorizing methods. However, it was noted post hoc that German Shepherds were at much higher risk of having MDR E. coli colonization on admission than other breeds (OR 6·20, 95% CI 1·33–28·93, P=0·020). The risk of being colonized with MDR E. coli on admission was lower for dogs with an underlying non-infectious medical disease or condition (OR 0·32, 95% CI 0·11–0·99, P=0·048) (diabetes mellitus, seizures, heart failure, autoimmune haemolytic anaemia and others) compared to those with an infectious disease or condition (dental disease, otitis externa, urinary infections and others). Dogs which had been referred were at a higher risk of having MDR E. coli colonization on admission than primary-care dogs (OR 1·94, 95% CI 1·03–3·65, P=0·040). Prior consultation and/or hospitalization (OR 2·66, 95% CI 1·28–5·53, P=0·009) and prior antimicrobial treatment (OR 5·14, 95% CI 2·58–10·25, P<0·001) were also associated with increased risk of MDR E. coli colonization on admission.
Of study dogs that had been hospitalized in the pre-admission period (68 cases, 53 controls), increasing days hospitalized in the pre-admission period was associated with a markedly increased risk of being colonized with MDR E. coli on admission. A number of other variables were also associated with an increased risk of MDR E. coli colonization on admission, including the number of times the dog was hospitalized, hospitalization at both the UQVTH and other hospitals, hospitalization in the large dog ward, ICU or chemotherapy ward at the UQVTH, duration and number of time of times a dog was catheterized and given intravenous (i.v.) fluids, interval between final i.v. fluid treatment and admission, general anaesthesia, surgery, diagnostic imaging techniques, other diagnostic tests, treatment with opioids and blood collection.
Of study dogs that received antimicrobials during the pre-admission period (47 cases, 17 controls), a number of antimicrobial-specific risk factors were associated with rectal colonization, including duration of treatment with any antimicrobials, the number of antimicrobials used, duration of treatment with amoxicillin/clavulanic acid, and antimicrobials given while in hospital or at home. Across all admissions, relative to those where no antimicrobial agents were used in the pre-admission period, odds of colonization at admission were much higher following treatment with fluoroquinolones (with or without one or more additional antimicrobials) (OR 24·85, 95% CI 3·8–∞, P<0·001) and were also elevated in admissions treated with non-fluoroquinolone antimicrobials (OR 4·00, 95% CI 1·89–8·76, P<0·001).
For the two risk factors for which the OR changed by >30% (interval between the final dose of any antimicrobial and admission and duration of prior amoxicillin/clavulanic acid treatment) the OR was further from 1 after adjusting for admission month, resulting in stronger associations between these risk factors and colonization with MDR E. coli on admission. However, after fitting with the final maximum-likelihood model neither were associated with MDR E. coli colonization on admission to hospital (P=0·477 and 0·423, respectively).
As infection control procedures at the UQVTH were in place during the study, we compared odds of being colonized for admissions that had been hospitalized in the first and second halves of the study period, with adjustment for admission month. The odds of being colonized did not differ significantly between dogs previously hospitalized at the UQVTH in the first and second halves of the study period, although the point estimate was consistent with lower odds in the second half (OR 0·84, 95% CI 0·22–3·10, P=0·538).
Multivariable analyses
The final maximum-likelihood multivariable model consisted of prior antimicrobial treatment, prior diagnostic imaging techniques and prior duration of hospitalization (Table 2). Only prior duration of hospitalization and prior diagnostic imaging techniques were associated with MDR E. coli colonization on admission to hospital. Using exact multivariable analysis, and after adjusting for days in hospital and diagnostic imaging techniques in dogs that had been hospitalized in the pre-admission period, prior treatment with fluoroquinolones (adjusted for prior use of other antimicrobial/s) was associated with increased risk of colonization on admission (OR 7·35, 95% CI 0·97–∞, P=0·054). After adjusting for prior fluoroquinolone treatment, admissions previously treated with non-fluoroquinolone antimicrobials (β-lactams, β-lactam/β-lactamase inhibitors, trimethoprim/sulfonamides and metronidazole) were not more likely to be colonized compared to previously untreated dogs (OR 0·87, 95% CI 0·25–2·78, P=1·000).
Table 2. Results of the final maximum-likelihood logistic model for risk factors for rectal colonization with MDR E. coli on admission to hospital, for 121 dogs (68 cases, 53 controls) in a retrospective, prevalence-based case-control study that had been hospitalized in the 42 days prior to admission

OR, Odds ratio; CI, confidence interval.
a Antimicrobials included amoxicillin/clavulanic acid, ampicillin, first generation cephalosporin, orbifloxacin, enrofloxacin, gentamicin, imipenem, clindamycin, metronidazole, doxycycline and trimethoprim/sulphonamide.
b Cumulative days hospitalized in the 42 days prior to the study admission.
c Diagnostic imaging techniques included radiography, echocardiography, ultrasonography and computer tomography.
Model fit and discriminatory ability
The observed numbers of cases and controls were close to that expected based on the final maximum-likelihood model. The Hosmer–Lemeshow goodness-of-fit test P value was 0·820. The discriminatory ability of the final model was moderate with the area under the ROC curve equal to 0·82. At a probability cut point of 0·52, the model's sensitivity and specificity were both about 0·74. These results indicate that, while the final model fitted the data, additional variables also determine whether dogs are colonized on admission.
Characterization of MDR isolates
The antimicrobial disk susceptibility profile and putative clonal groups of the 58 isolates that were characterized are shown in Table 3. Nineteen of the isolates were identified via multiplex PCR as putative CG1 strains (positive for uspA, dfrA17-aadA5 and bla CMY) and 36 isolates as CG2 strains (positive for uspA and bla CMY only). Three isolates could not be placed into either clonal group. These isolates were all identified as E. coli; two contained the dfrA17-aadA5 gene and were possibly CG1 strains that had lost bla CMY, whereas the remaining isolate may have been a CG2 strain that had also lost bla CMY, a CG1 strain that had lost both the integron and bla CMY, or an unrelated isolate.
Table 3. Putative clonal groupFootnote a and resistance profiles for MDR E. coli isolates from 82 case (colonized) dogs on admission to The University of Queensland Veterinary Teaching Hospital between 1 March 2001 and 30 October 2002
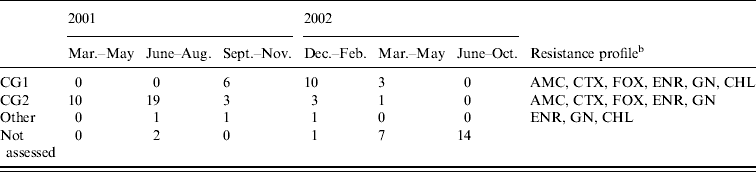
a Clonal group 1; positive for E. coli uspA, bla CMY and dfra17-aadA5 and clonal group 2; positive for uspA and bla CMY only, Other; all contained uspA, two contained dfrA17-aadA5 [Reference Sidjabat14, Reference Sidjabat15].
b AMC, amoxicillin/clavulanic acid; CTX, cefotaxime; FOX, cefoxitin; ENR, enrofloxacin; CHL, chloramphenicol; GEN, gentamicin.
DISCUSSION
These results confirm that duration of previous hospitalization is an important risk factor for dogs being colonized with MDR E. coli on admission to a large veterinary teaching hospital. This is supported by other studies conducted in ICUs which show that the proportion of canine patients with resistant rectal E. coli increases with the duration of hospitalization [Reference Filius7, Reference Yagci8, Reference Ogeer-Gyles11]. People hospitalized in ICUs for ⩾7 days are 2–3 times more likely to be colonized with an antimicrobial-resistant pathogen than those hospitalized in ICUs for shorter periods [Reference Fridkin19]. While prolonged hospitalization may also be associated with more severe underlying disease and/or immunocompromised patients, we speculate that dogs with prolonged hospitalization periods have more opportunity to be exposed to the hospital environment, including other hospitalized animals and hospital staff.
Environmental reservoirs of bacteria in veterinary hospitals contribute to the occurrence of nosocomial infections [Reference Eugster20, Reference Boerlin21]. At the UQVTH, certain areas of the hospital (ICU, small and large dog ward, and the food preparation area) were heavily contaminated and were therefore a reservoir of MDR E. coli during the early phases of the infection control programme [Reference Sidjabat14]. MDR E. coli was rarely isolated from the hospital environment after May 2001 following implementation of new infection control procedures (M. Honnery, personal communication). However the risk of dogs being colonized with MDR E. coli was not greatly different between those previously hospitalized at UQVTH in the first and second halves of the study period. The gastrointestinal tract is the other major reservoir of MDR E. coli and it is possible that MDR E. coli sub-populations persist undetected in the other commensal coliforms within the gut, only to proliferate in response to antimicrobial selection pressure.
Individual antimicrobial treatments were difficult to evaluate as there were many antimicrobial combinations prescribed in the pre-admission period and some antimicrobials were only given to a small number of dogs. This may have resulted in failure to identify a relationship between particular antimicrobial agents and MDR E. coli colonization. On univariable analysis, prior antimicrobial treatment appeared to be significant, but when duration of hospitalization and diagnostic imaging techniques were accounted for in multivariable analysis, fluoroquinolones were the only antimicrobial agent associated with MDR E. coli colonization. Although the confidence interval for the odds ratio was wide and included values <1, the point estimate was high (7·35) and this result was readily compatible with a large effect. Previous evidence also supports the hypothesis that fluoroquinolone antimicrobials are a risk factor for MDR E. coli colonization [Reference Yagci8, Reference Ogeer-Gyles11].
Prior use of fluoroquinolones was also identified as a possible risk factor for carriage of fluoroquinolone-resistant E. coli in dogs hospitalized in an ICU in a previous study [Reference Ogeer-Gyles11]. Prior fluoroquinolone treatment has also been identified as a risk factor for colonization with fluoroquinolone-resistant Gram-negative bacilli in humans before admission to hospital and/or receipt of new therapy [Reference Yagci8]. Prior exposure to fluoroquinolones has been associated with quinolone-resistant E. coli bacteraemia [Reference Garau22], urinary tract infections [Reference Lin23] and nosocomial infections [Reference Richard24] in humans. Dogs treated with fluoroquinolones shed substantially higher population densities of MDR E. coli for longer durations compared to untreated dogs [Reference Trott16].
An interesting finding from this study was that neither β-lactam nor potentiated β-lactam antimicrobials appeared to increase the risk of colonization with MDR E. coli on admission to hospital. This was unexpected as all but three of the MDR E. coli isolates characterized contained an AmpC β-lactamase. We expected that treatment with these antimicrobials would be a risk factor due to the selection pressure required for maintenance of the plasmid. Further research is therefore required to investigate this finding.
Prior diagnostic imaging techniques were also a risk factor for MDR E. coli colonization on admission to hospital. We suspect that this reflects increased handling or movement of the animal within hospitals prior to the study admission and/or greater severity of prior illness. The ultrasonography, radiography and computer tomography rooms at the UQVTH did not yield a positive environmental swab for MDR coliforms during the infection control programme [Reference Sidjabat14]. Other studies examining risk factors for gastrointestinal colonization of Enterobacteriaceae in humans have identified similar risk factors such as urinary catheterization and mechanical ventilation [Reference Lucet25].
The current study has a number of limitations. Exposure variables were only examined for 42 days prior to admission and it is possible that exposures prior to this may have influenced colonization. It was not possible to swab every dog on admission due to staff availability. However, it is unlikely that this was associated with the probability of being colonized or with exposure status. The selective media (MCAEG) used may have prevented some MDR E. coli from being detected, although this was probably uncommon given that the resistance profile of isolates characterized from rectal swabs corresponded to those of the clinical isolates obtained from extraintestinal infections. Rectal swabbing was undertaken by different people (staff and students) and it was possible that this led to some sample variability. Previous studies indicate that while rectal swabbing is highly specific for identifying gastrointestinal carriers, some colonized dogs may not be detected [Reference Ogeer-Gyles11, Reference Lautenbach26]. Case-control studies are not ideal for assessing rare exposures, and, for some variables, we had to collapse some categories into broad groups to ensure models converged and to obtain moderately precise effect estimates.
A number of other variables which on univariable analysis appeared important were excluded during the multivariable modelling process. These included: underlying infectious disease or condition; previous hospitalization in the large dog ward or ICU at the UQVTH; prior treatment with i.v. fluids, steroids and NSAIDs; and prior anaesthesia, and surgical and diagnostic procedures. Some of these variables may be important risk factors for dogs being colonized with MDR E. coli on admission, but in this study, they failed to be independently significant after accounting for duration of hospitalization and diagnostic imaging techniques.
In conclusion, these results show that dogs hospitalized for ⩾4 days and those treated with fluoroquinolones or undergoing diagnostic imaging techniques in the 42 days prior to admission are at increased risk of being colonized with MDR E. coli on admission to hospital. The study did not aim to identify whether these risk factors increase the incidence of acquiring colonization or the duration of colonization once acquired. However, we would expect that prior antimicrobial treatment would increase both [Reference Donskey2], whereas duration of hospitalization and diagnostic imaging techniques would be expected to increase the incidence of acquiring MDR infection. As animals rectally colonized with MDR E. coli on admission to hospital are at greater risk of acquiring MDR E. coli extraintestinal infections, additional precautions could be taken to prevent extraintestinal nosocomial infections in these risk groups as has been proposed for colonized humans. Animals in the risk groups identified in this study could also be more closely monitored and/or managed according to infection control guidelines and possibly isolated from the general hospital and outpatient populations to limit the potential for transmission of MDR E. coli between hospitalized dogs and from these dogs to hospital personnel.
NOTE
Supplementary material accompanies this paper on the Journal's website (http://journals.cambridge.org/hyg).
ACKNOWLEDGEMENTS
We thank the staff and students at UQVTH for the collection of rectal swabs. We also acknowledge The University of Queensland Veterinary Diagnostic Laboratory for the collection and storage of isolates, in particular Susan Moss and Dr Kirsty Townsend.
DECLARATION OF INTEREST
None.





