INTRODUCTION
Diarrhoeal disease is a major public health problem in developing countries and is considered the foremost cause of death [Reference Guerrant1–Reference Amisano4] accounting for about 2 million deaths worldwide annually [Reference Kosek, Bern and Guerrant5–Reference Bryce6], afflicting persons of all age groups and status [Reference Caprioli7]. The major source of the disease is the poor quality of accessible drinking water, contaminated food, poor standard of personal hygiene and lack of appropriate sanitation. It is noteworthy that the aetiological agents of diarrhoea vary with localities within an environment, socioeconomic status and cultural practices [Reference Cairncross8]. Therefore, knowledge of the aetiological agents is arguably important for adequate address of epidemiological surveillance and proper treatment. There is a tight correlation between diarrhoeal diseases and lack of adequate water worldwide [Reference Momba, Madoroba, Obi and Mendez-Vitas9]. In many developing countries, there is improper waste management including the handling of faecal materials. The outcome of this is the eventual leaching of such waste matters into canals and well water through the aid of flooding. There is a wide spectrum of microbes, including bacteria, viruses and parasites that have been implicated, either individually or synergistically, in the aetiology of the disease [Reference Ochoa, Salazar-Lindo and Cleary10–Reference O'Ryan, Prado and Pickering12]. The bacterial agents of diarrhoeal disease are commonly Escherichia coli, Shigella spp., Salmonella spp., Campylobacter, Yersinia, Aeromonas, etc. [Reference Presterl13]. Individuals afflicted with these bacteria in developing countries benefit from antibiotic therapy often via self-medication, since the antibiotics are commonly available without prescription. However, non-compliance with regard to dosing requirements is an issue due to the cost of the antibiotics [Reference Okeke, Lamikanra and Edelman14]. There is a consequent progressive increase in antibiotic resistance in enteric pathogens in developing countries in a manner that poses a critical public health threat [Reference Blake15–Reference Chigor17].
The current approach is to ascertain that the pathogen of interest is identified using culture- and non-culture-based molecular techniques with sometimes protracted turnaround times. This issue of time is further compounded by the wide diversity of bacterial and viral aetiological agents of diarrhoeal disease [Reference Girard18], thus complicating accurate diagnosis and necessary surveillance in a developing country. The scenario, therefore, is that of a disease burden that could be easily addressed on a long-term basis with the provision of portable clean water, better sanitation, proper hygiene and improved nutritional practices. Immediate efforts directed at addressing the disease burden itself are hampered by lack of proper diagnosis and abuse of antibiotics.
Real-time detection of multiple aetiological agents of diarrhoea from readily available specimens, such as stool, would contribute immensely to efficient clinical and public health responses to diarrhoea outbreaks, cases of chronic diarrhoea and epidemiological surveillance. An important development in the capacity to detect multiple aetiological agents is the use of such versatile methods used in the detection of toxins elaborated by bacterial agents of diarrhoea. For instance, enterotoxigenic E. coli produces one or more heat labile toxins (LT1 and LT2); it also produces heat stabile toxins (STa and STb), which are of extreme importance as causal virulence factors of diarrhoea [Reference Flores19–Reference Zhang21]. The identification and evaluation of stool pathogens in a highly populated and large area such as Lagos metropolis in Nigeria will go a long way in providing much needed information on the preponderance of a pathogen among stool pathogens and reveal the relationship between pathogenicity and various strains of the most implicated pathogen. On this basis, we decided to adopt Lagos as a typical highly urbanized city in a developing country and investigate the breadth of enteropathogen profiles in stool in a manner that reduces the turnaround time and determines the links between toxigenic strains of E. coli being the most frequently found bacteria of stool and accessible drinking water. The findings on E. coli toxin genes were compared with similarly investigated samples in Wisconsin, USA as a validation control; with the expectation that a pool of data that will be beneficial to the public health in the city will be assembled. Moreover, since the therapeutic interventions in Lagos sometimes involve indiscriminate use of antibiotics which predisposes antibiotic resistance, we decided to investigate if there is an observable pattern in the relationship between antibiotic-resistant E. coli and toxin elaboration; thus revealing an additional burden on the public health of the residents of such large city.
METHODS
Materials
All chemicals and reagents were purchased from Sigma-Aldrich Chemical Company (St Louis, USA) unless otherwise stated.
Study population and samples
The study population consisted of adult patients (aged 17–57 years) from seven localities (Alimonsho, Shomolu, Oshodi, Surulere, Yaba, Ikeja, Mushin) in Lagos, Nigeria, who were attending hospital with complaints about diarrhoeal disease and consented to provide stool samples. A total of 102 stool samples were obtained: 91 chronically ill diarrhoea patients and 11 controls from healthy volunteers. Further, 25 acutely ill diarrhoea patients' samples from Milwaukee and Racine in Wisconsin state, USA were collected from consenting individuals (18–55 years). Neither patients nor controls admitted to having received antibiotic treatment within 2 weeks before submission of samples. To relate findings in stool samples with accessible water in the same localities, we collected 117 water samples from seven localities in Lagos as described above from different sources: canals, wells, sachet packed water and municipal pipe-borne ‘tap’ water. Also, from Milwaukee and Racine in Wisconsin state, USA, 26 water samples from similar sources, i.e. local beaches, storm drains and water treatment plant were collected. All samples (stool and water) were aseptically collected.
Preparation of sample and culture of Escherichia coli
The isolation of E. coli involved the inoculation of fresh stool samples onto Stuart's transport medium (Oxoid, UK), maintained on ice during transportation to the laboratory for same day processing. The water samples were transported in sterile containers on ice to the laboratory and processed in the same manner as the stool samples. The samples were inoculated into peptone water (Oxoid) and incubated at 37°C for 18 h. The samples were subsequently streaked onto Eosin Methylene Blue (EMB) agar (Oxoid) and incubated overnight at 37°C. Indole, Methyl Red, Voges–Proskauer reaction and Simmons' citrate (IMViC) tests were performed on the colonies that showed E. coli growth characteristics on EMB agar. Analytical profile index (API) 20E strips (bioMérieux, France) were also used to confirm the identity of isolates as E. coli [Reference Bebora22, Reference Kariuki23].
Susceptibility studies
The E. coli isolates from Lagos were exposed to 16 antibiotics (ciprofloxacin 5 μg, trimethoprim/sulfamethoxazole 1·25/23·75 μg, ceftazidime 30 μg, streptomycin 10 μg, nitrofurantoin 300 μg, ofloxacin 5 μg, amoxycillin/clavulanate potassium 20 μg/10 μg, gentamicin 10 μg, cefaclor 30 μg, ampicillin 10 μg, norfloxacin 10 μg, tetracycline 30 μg, cefuroxime sodium 30 μg, nalidixic acid 30 μg, ceftriaxone 30 μg, cefixime 30 μg) reflecting different classes of antibiotics. This enabled the capture of the susceptibility pattern using disk diffusion on Mueller–Hinton agar (MHA) plates. All the E. coli strains showing resistance to the various antibiotics used in the susceptibility studies were further selected for virulence toxin detection using conventional multiplex real-time PCR described below.
Luminex xTag Gastrointestinal Pathogen Panel (GPP) procedure (Luminex Corp., USA)
The profiling of enteropathogens was conducted using the Luminex xTag Gastroplex (GPP) Analyte Specific Reagent (ASR) [Reference Navidad24], designed to detect 16 targets of various pathogens and their toxin genes that are associated with human gastroenteritis. An extra target for detecting the internal control and identifying PCR inhibitory specimens (bacteriophage MS2), was added to the sample prior to extraction. Targets are RNA and DNA of viruses, bacteria and parasites. In this instance, the target pathogens were adenovirus 40/41, Campylobacter spp., Clostridium difficile toxin A/B, Cryptosporidum spp., Entamoeba histolytica, Escherichia coli O157, enterotoxigenic Escherichia coli LT/ST, Giardia spp., norovirus G1/G11, rotavirus A, Salmonella spp., shiga-like toxin producing Escherichia coli, Shigella spp., Vibrio cholera, Yersinia enterocolitica. The method essentially is a modification of multiplex RT–PCR with Luminex's proprietary universal Tag sorting system on the Luminex xMAP platform. Prior to the GPP procedure, DNA was extracted from stool samples using bioMérieux NucleSense easy MAG extraction system according to the manufacturer’ instructions. In brief, 20 μl PCR mix and 5 μl DNA template was added to each tube, then the mixture was vortexed and placed in a thermal cycler. After pre-heating at 53°C, DNA extracts were loaded and the run was started. Seventy-five microlitres of hybridization mix was added to each well; the plate was sealed and returned to the thermal cycler. The GPP analysis was performed using the Bio-Plex®−200 system (Bio-Rad Laboratories, USA) and Luminex xPONENT® software (Luminex Corp.).
Conventional multiplex using real-time PCR
The elaboration of virulence toxin genes was determined using conventional multiplex real-time PCR. In brief, this incorporated 22 primers for the amplification of 11 virulence genes (stx1, stx2, eae, bfp, LT, st11, virF, ipaH, aaFII, daaE, uidA) exhibited by diarrhoeagenic E. coli [Reference Carey25].
Statistical method
The expression of toxin genes by multiple antibiotic-resistant E. coli was analysed by one-way analysis of variance (ANOVA) followed by Bonferroni's post-hoc test, using GraphPad Prism 5.01 software (GraphPad Software Inc., USA). The level of statistical difference was set at P ⩽ 0·05.
RESULTS
Assessment of the enteric pathogen profile in stool of diarrhoea patients in Lagos, Nigeria
The simultaneous detection of 16 gastrointestinal pathogens in 91 diarrhoeal stool samples of adult patients in Lagos showed that 12 (75%) out of 16 targeted pathogens were detected. All 11 control stool samples from healthy individuals were negative for the targeted pathogens except E. coli which were detected in two samples. Figure 1 represents the detection rate of pathogens. Ent. histolytica was the most frequently detected pathogen (19%), followed by enterotoxigenic E. coli represented as E. coli that expresses heat labile toxin genes (LT) and heat stabile toxin genes (ST) totalling (14%). Campylobacter, detected once, was the least detected pathogen. Three viruses were detected: norovirus G1/G11 (8%), adenovirus 40/41 (5%), and rotavirus A (3%). Out of 91 stool samples, 50 contained pathogens of various numbers, 31 samples had one pathogen, while 19 samples contained two or more pathogens.

Fig. 1. Percentage of pathogens or virulence toxin genes in adult diarrhoeal stool samples collected from patients in various localities in Lagos, Nigeria. This depicts the simultaneous detection of 16 pathogens using the Luminex xTAG Gastro-intestinal Pathogen Panel (GPP). This figure represents the total of each pathogen type in the whole of Lagos metropolis in Nigeria.
Simultaneous determination of the enteric pathogen profile in stool of diarrhoea patients from individual localities in Lagos metropolis
The stool pathogen profile in seven localities in Lagos metropolis was individually assessed. Alimonsho stool samples revealed a pathogen profile pattern in which Campylobacter was the most common pathogen (Fig. 2a ), while in the Shomolu area, E. coli (E. coli LT and ST taken together) and Giardia were the most common pathogens detected (Fig. 2b ). The detection of pathogens in the Oshodi locality (Fig. 2 c) was very sparse. In instances where a pathogen was detected, only one sample was positive. The reason for the Oshodi data is unclear, but it is noteworthy that cost related non-compliance with dosing may not be common in Oshodi, where most inhabitants being traders can afford self-medication cost. In Surulere (Fig. 2 d), Ent. histolytica and E. coli (LT and ST) were detected along with Salmonella, adenovirus and norovirus. In other localities (Yaba, Ikeja, Mushin; Fig. 2 e–g, respectively), the pattern was one in which E. coli ST (Fig. 2 e), or LT (Fig. 2 e–g), or shiga 2 (Fig. 2 e, f) were detected, although in scanty proportions.

Fig. 2. Percentage of pathogens or virulent toxin genes in adult diarrhoeal stool samples. The stool pathogen profile from diarrhoea patients from individual localities in Lagos, Nigeria is shown: (a) Alimonsho; (b) Shomolu; (c) Oshodi; (d) Surulere; (e) Yaba; (f) Ikeja; (g) Mushin.
Detection of toxin genes expressed by E. coli isolated from accessible drinking-water sources in Lagos and Wisconsin, USA
Arguably, the frequency of detection of E. coli and some of its associated toxins during stool pathogen profiling in Figures 1 and 2, may reasonably be a reflection of a the relationship between toxigenic E. coli distribution in accessible drinking water and the clinical disease of diarrhoea; therefore, we decided to examine the expression of E. coli toxin genes in accessible drinking-water sources in Lagos and compare it with similar water sources from Wisconsin, although not usually ingested water sources, since the accessible drinking water in Wisconsin is better treated. Results (Fig. 3 a, b) show that Lagos isolates expressed toxin genes [eae (42%), LT (10%), stx2 (6%), bfp (5%), stx1 (4%), ST11 (2%), virF (1%), aaFII (1%), daaE (1%)] of biological significance, while Wisconsin water isolates (although not usual drinking water) expressed higher levels of stx1 (14%) and stx2 (25%) toxin genes which may be of biological significance; but lesser levels of eae (35%) and LT (2%). The biomolecule uidA is characteristically expressed in E. coli.

Fig. 3. Virulence toxin genes expressed by Escherichia coli isolates obtained from water sources. (a) The detection of toxin genes of E. coli isolates from accessible drinking water (canal, well, municipal pipe borne and sachet packed water) in Lagos, Nigeria. (b) Toxin genes of E. coli detected in water sources (beach, storm and treatment plant water) in Wisconsin, USA.
Detection of toxins elaborated by E. coli isolated from adult diarrhoeal stool samples in Lagos and Wisconsin
Based on the finding that E. coli isolates from accessible drinking-water sources in Lagos elaborated toxins of concern in diarrhoea, we decided to investigate the possible elaboration of the same profile of toxins in diarrhoeal stool samples from Lagos and determine if our findings could represent a pattern illustrating a correlation between toxins of accessible drinking sources and those of diarrhoeal disease sources. Results (Fig. 4 a) show an expression of toxin genes [eae (78%), LT (18%), stx2 (3%), bfp (11%), stx1 (3%), ST11 (4%), daaE (2%)] from E. coli isolates which may be related to those from accessible drinking-water sources (Fig. 3 a). To determine if this pattern of relatedness between water sources and diarrhoeal disease sources can be replicated in Wisconsin, we assessed the expression of toxin genes by E. coli isolates from diarrhoeal stool samples in Wisconsin (Fig. 4 b). While E. coli isolates from water in Wisconsin averagely expressed toxin genes eae, a higher expression was obtained in stool (54%) and LT was unexpressed (Fig. 3 b). Wisconsin stool isolates expressed significantly high levels of stx2 (50%), and stx1 (27%), suggestive of the fact that diarrhoeal disease in Wisconsin may be related to water to an appreciable extent.

Fig. 4. Virulence toxin genes of Escherichia coli isolated from stool strains. The detection of toxin genes expressed by E. coli isolated from adult diarrhoeal stool in Lagos metropolis in (a) Nigeria and (b) Wisconsin, USA is shown.
Pattern of virulence toxin gene expression by diarrhoeagenic E. coli strains that are resistant to various antibiotics
In order to investigate if the antibiotic susceptibility pattern of E. coli isolates bear any relationship with toxin elaboration, we decided to determine toxin gene expression by antibiotic-resistant diarrhoeagenic E. coli. Table 1 shows the resistance patterns to 16 antibiotics and the expression of toxin genes by E. coli strains from diarrhoeal stool samples, while Figure 5 shows the expression of E. coli toxin genes and the extent (%) of resistance to multiple antibiotics. The stx1 toxin gene was detected in strains resistant to 13 (85%) of the antibiotics used. It was not elaborated by isolates resistant to ceftriaxone, cefixime and gentamycin. The stx2 gene was expressed by isolates resistant to 11 (69%) of the antibiotics used and was not expressed by isolates resistant to ciprofloxacin, ofloxacin, norfloxacin, ceftriaxone and gentamycin. The eae toxin was identified in strains resistant to 15 (93·75%) antibiotics used. It was not detected in nitrofurantoin resistance. The bfp toxin gene was identified in isolates resistant to seven (43·75%) antibiotics. It was not detected in those resistant to floroquinolones (ciprofloxacin, ofloxacin, norfloxacin), nalidixic acid, ceftriaxone, cefuroxim, ceftazidime, nitrofurantoin and amoxcillin/clavulanic acid. The LT toxin gene was detected in isolates resistant to all (100%) the antibiotics used. The ST11 toxin gene was detected in isolates resistant to six (37·50%) antibiotics. It was not detected in those resistant to cephalosporins (ceftriaxone, cefixime, cefuroxime, ceftazidime, except cefaclor), ciprofloxacin, nalidixic acid, amoxycillin/clavulanic acid, nitrofurantoin and gentamycin. The virF toxin gene was found in the strains resistant to four (25%) antibiotics. It was not identified in those resistant to floroquinolones (ciprofloxacin, ofloxacin, norfloxacin), the cephalosporins (ceftriaxone, cefixime, cefuroxime, ceftazidime, cefaclor), nalidixic acid, nitrofurantoin, amoxycillin/clavulanic acid, and gentamycin. The aaFII toxin gene was detected in isolates resistant to three (18·75%) antibiotics. It was not detected in those resistant to floroquinolones (ciprofloxacin, ofloxacin norfloxacin), cephalosporins (ceftriaxone, cefixime, cefuroxime, ceftazidime, except cefaclor), nalidixic acid, tetracycline, streptomycin, fluoroquinolones, nitrofurantoin, amoxycillin/clavulanic acid, gentamycin. The daaE toxin gene was detected in isolates resistant to eight antibiotics (50%). There was no daaE toxin indicated in isolates resistant to ceftriaxone, tetracycline, cefuroxime, ceftazidime, streptomycin, nitrofurantoin, amoxicillin/clavulanic acid, and gentamycin.

Fig. 5. Toxin gene expression by Escherichia coli and the extent (%) of antibiotic resistance exhibited. The E. coli isolates that expressed stx2 genes exhibited a significantly lower percentage of multiple antibiotic resistance compared to those that expressed stx1, or eae, or LT toxin genes. * P ⩽ 0·0001.
Table 1. The expression of virulence toxin genes by antibiotic-resistant diarrhoeagenic Escherichia coli from stool samples in Lagos, Nigeria
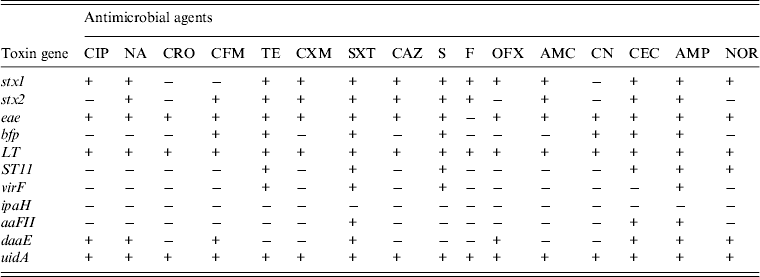
+, Resistant; −, sensitive.
The antibiotics assessed were ciprofloxacin (CIP); nalidixic acid (NA); ceftriaxone (CRO); cefixim (CFM); tetracycline (TE); cefuroxime (CXM); sulfamethoxazole/trimethoprim (SXT); ceftazidime (CAZ); streptomycin (S); nitrofurantoin (F); ofloxacin (OFX); amoxicillin/clavulanic acid (AMC); gentamicin (CN); cefaclor (CEC); ampicillin (AMP), norfloxacin (NOR).
The toxin gene expression assessed were shiga-like toxin (stx1 and stx2), intimin-E. coli attaching and effacing (eae), bundle forming pilus (bfp), heat labile toxin (LT), heat stabile toxin 11 (ST11), transcriptional activator virulence gene (virF), invasive plasmid antigen H (ipaH), aggregative adherence fimbriae II (aaFII), diffuse adherent (daaE), β-glucuronidase (uidA).
DISCUSSION
It is increasingly obvious that a wide array of bacterial, viral and parasitic pathogens are capable of infecting the intestine with or without overt dehydrating diarrhoea burden. Although diarrhoeal disease is worldwide in distribution, developing countries are more impacted by the disease burden. The rate of mortality in many countries is on the decline while the morbidity rate remains high [Reference Kosek, Bern and Guerrant5]. Arguably, the morbidity burden of diarrhoea is accounted for by the systematic malnutrition caused by persistent diarrhoea and chronic enteropathy arising from recurring enteric infections. In developing countries, the problem of diarrhoea is related to poor sanitary conditions as observed in the standard of hygiene in food and water consumed that are often contaminated with an array of enteric pathogens. Thus, 1/6 people (1·1 billion individuals) have no source of safe water and 4/10 (2·6 billion individuals) lack even pit latrines (which on its own is a source of pathogen distribution during flooding), which results in numerous enteric infections and worsening the rates of morbidity from diarrhoea [Reference Mara26]. The burden of diarrhoeal disease on the developing world is tremendous when the malnutrition effect on infants and children is considered. Furthermore, infection with specific enteric pathogens such as enteroaggregative E. coli (EAEC) and Cryptosporidium spp. can negatively impact growth even in the absence of overt diarrhoea [Reference Checkley27–Reference Steiner30].
Early detection of the wide array of pathogens involved in diarrhoea would contribute to intervention and control strategies. There are points of intervention that can lead to the annulment of the cycle of diarrhoea and its burden; these include the advent of new molecular probes that increasingly reveal important viral, bacterial, and parasitic enteric pathogens and their virulence traits and the addressing of the long-term impact of diarrhoea on development and productivity. The use of the Luminex xTAG GPP, enabled multiplex detection of multiple gastrointestinal pathogens and toxins simultaneously in one stool sample as depicted in Figure 1. In this way, the role of multiple infections in the complication of diarrhoeal disease and the impact on disease severity is revealed. The use of xTAG GPP has gone a long way in showing that the gastrointestinal pathogenic burden in a complex city such as Lagos can be analysed creating room for effective infection control and proper antimicrobial therapy. The xTAG GPP is a variant of multiplex which incorporates culture independence, sensitivity and specificity thus having an enormous prospect as a screening tool in routine clinical testing and public health diarrhoea outbreak investigations. The importance of xTAG GPP becomes very obvious when one considers the fact that the current culture- and non-culture-based molecular methods have a much shorter turnaround time.
The detection of a wide range of pathogens in diarrhoeal stool samples from Lagos, Nigeria is important, and of special significance is the detection of some E. coli toxin genes in the xTAG GPP procedure (Figs 1, 2 a–g, respectively). Therefore, it was imperative to further investigate the genes expressed by E. coli isolates from stool and water sources (Figs 3 a, b, 4 a, b). The intent here is to determine the link between toxigenic strains of E. coli from stool and accessible drinking-water sources. The E. coli isolates from Lagos were found to express genes that could lead to elaboration of toxins that are highly pathogenic and thus, a serious threat to public health. The expression of the eae toxin gene, which encodes for intimin, an intestinal adherence factor, is widely distributed in Lagos. Of interest is the fact that eight diarrhoeagenic E. coli virulence toxin genes (uidA, eae, LT, bfp, stx2, stx1, ST11, daaE) were detected from the Lagos diarrhoeal stool samples (Figs 2 a–g, 4 a) while nine virulence toxin genes (uidA, eae, stx2, stx1, virF, aaFII, bfp, ST11, daaE) were detected in accessible drinking-water isolates (Fig. 3 a), thus the Lagos water isolates had more toxin genes expressed compared to the stool isolates.
The Wisconsin, USA water isolates expressed comparatively less eae and LT toxin genes (Fig. 3 b) and the stool isolates expressed significantly high levels of eae, stx2, stx1 toxin genes (Fig. 4 b) apart from the uidA toxin genes expressed in either cases. The emerging facts in the comparative assessment of the virulence toxin gene expression between Lagos and Wisconsin isolates are that stx1 and stx2 toxins (associated with enterohaemorrhagic E. coli activities) were expressed more frequently in Wisconsin water isolates than Lagos, a fact which may not be unconnected with the water sources. It is noteworthy that despite the similarities between the Wisconsin and Lagos water sources studied, in Wisconsin, water from these sources are not drinking water, although it is arguable that they may be accidentally consumed. Moreover, it is reasonable to assume that for the purpose of consumption, the Lagos sources may be subjected to relative filtration which may not significantly affect bacteria load; therefore the presence of enterohaemorrhagic E. coli toxins may be more impactful in Lagos than Wisconsin. The high levels of toxin gene (stx1 and stx2) expression in Wisconsin diarrhoeal stool points to the fact that samples were obtained from acutely ill diarrhoea patients compared to Lagos where patients had chronic diarrhoea. Taken together, the Lagos studies show relatedness between water as a pathogen source and the onset of clinical diarrhoea.
There is compelling evidence of the indiscriminate use of antibiotics in the city of Lagos; therefore, there is predisposition of antibiotic resistance. The public health implication of this, alongside the finding of extensive toxin gene expression by E. coli isolates both in environmental and stool samples is that an additional burden is borne by patients afflicted by antibiotic-resistant E. coli which also expresses virulence toxin genes. This is significant, since among the bacterial agents of diarrhoeal aetiology, diarrhoeagenic E. coli occupies a central place. It is therefore imperative that the combination of two burdens (antibiotic resistance and toxin elaboration) calls for strict public health measures that will ensure discipline in therapeutic interventions that involve the use of antibiotics. For instance, it was observed that eae, LT, and stx1 and stx2 (virulence factors of shiga toxin-producing E. coli), were the most preponderant toxin genes expressed by antibiotic-resistant Lagos isolates (Table 1). Arguably, more severe cases of diarrhoea will be evident in those localities where the combined burden of drug resistance and toxin elaboration co-exist. Insights into the statistical relevance of multiple antibiotic-resistant E. coli isolates to eae, LT, and stx1 and stx2 toxin gene expression (Fig. 5), show the following toxin gene expression pattern stx2 < stx1 < eae < LT. Although the approach reflecting the combined burden of antibiotic resistance and elaboration of virulence toxin by antibiotic-resistant E. coli on public health is a unique outlook, there is evidence suggesting that these factors are of formidable pathogenesis concern. For instance, the capacity to elaborate shiga toxins is a unique pathogenesis asset to E. coli and has an important role in causing disease which may be life-threatening, especially if the disease is accompanied by bloody diarrhoea and haemolytic uraemic syndrome [Reference Checkley29, Reference Steiner30]. Indeed, the global significance of poor sanitation-based dissemination of infection in low-income countries should not be underestimated. The importance of this can be seen in relation to horizontal gene transfer of virulence factors resulting in the emergence of new pathotypes as evident in the recent case of the shiga-toxin-producing EAEC pathotype thought to be derived from sprouted seeds from Egypt which were involved in a large uraemic outbreak in Germany and Europe in 2011 [Reference Griffin, Karmali and Goglio31–Reference Frank33].
ACKNOWLEDGEMENTS
This work was supported by funds from Milwaukee Health Department Laboratories, and funds made available in the form of the Shaw Scientist Award to Professor A. A. Azenabor by the Greater Milwaukee Foundation.
DECLARATION OF INTEREST
None.









