The International Committee on Taxonomy of Viruses (ICTV) has classified the Parvovirinae subfamily viruses into five genera: Dependovirus, Bocavirus, Erythrovirus, Parvovirus and Amdovirus. In general, the genome of a Parvovirinae virus is a linear, single-stranded DNA of about 5000 nucleotides (nt) containing two major open reading frames (ORFs) coding for the non-structural protein (ORF1) located at the 5′-end and the capsid protein (ORF2) located at the 3′-end. An additional ORF3 located in the middle of the viral genome is observed in members of the Bocavirus genus. The ICTV taxonomy (2009 release) listed two viral species, bovine parvovirus (BPV1) and canine minute virus (CMV), belonging to the Bocavirus genus. BPV1 was reported in 1961 [Reference Abinanti and Warfield1] and the genome sequence was reported in 1986 [Reference Chen2]. CMV was isolated in 1967 [Reference Binn3] and the genome sequence was reported in 2002 [Reference Schwartz4]. Since 2005, new bocavirus or boca-like viruses (i.e. having an additional ORF3) have been identified in other animal species. The first human bocavirus (HBoV1) was reported in 2005 [Reference Allander5] and HBoV2, HBoV3 and HBoV4 were identified in 2009–2010 [Reference Kapoor6–Reference Kapoor8]. Gorilla bocavirus species 1 (GBoV1) was identified in 2010 [Reference Kapoor9]. Between 2009 and 2010, six different porcine bocaviruses originally named: porcine boca-like virus (PBoLV), porcine parvovirus 4 (PPV4), PBoV1, PBoV2, 6V and 7V, were reported. PBoLV was first identified in Sweden as a partial DNA clone of 1879 nt derived from the lymph nodes of animals with porcine circovirus-associated disease (PCVAD) in 2009 [Reference Blomström10] and subsequently detection of this virus in China expanded the genome sequence to 4786 nt in 2011 [Reference Zeng11]. PPV4 was first identified in the lung lavage of PCVAD animals in the USA as a head-to-tail circular or concatemeric molecule (5905 nt) [Reference Cheung12] and subsequently confirmed in China in 2010 [Reference Huang13]. PBoV1 (5173 nt), PBoV2 (5186 nt), 6V (2407 nt) and 7V (2434 nt) identified in faecal samples of healthy animals in China were reported in 2010 [Reference Cheng14]. The partial genome sequences of 6V and 7V contained portions of ORF3 and the complete ORF2 of the virus. It should be noted that the 5′- and 3′-termini sequences of porcine bocaviruses have not been determined; however, the almost full-length sequences available cover all three ORFs.
In this study, we first examined the genetic relationships of the porcine bocaviruses with those identified in other animal hosts and four representative viruses from humans. We then determined the prevalence of each porcine bocavirus in the swine population from ten Chinese provinces.
A total of 13 bocavirus and boca-like virus sequences were downloaded from NCBI: six from swine (PBoLV: HQ223038; PPV4: GQ387499; PBoV1: HM053693; PBoV2: HM053694; 6V: HM053672; 7V: HM053673), one from cattle (BHV1: NC_001540), one from dog (CMV: NC_004442), one from gorilla (GBoV1: HM145750), and four from humans (HBoV1: DQ000495; HBoV2: NC_012042; HBoV3: NC_012564; HBoV4: FJ973561). Only partial sequences were available for 6V and 7V that contained part of ORF3 and all of ORF2. The nucleotide sequences obtained were aligned and a phylogenetic tree was generated using the ORF2 deduced amino-acid sequences (Fig. 1) as previously described [Reference Huang13, Reference Zhai15]. The results showed that the primate and animal bocaviruses differ significantly and formed two separate clusters. Interestingly, the porcine bocaviruses exhibited much greater diversity than the primate viruses. Of the porcine bocaviruses, PBoV1 was closely related to PBoV2 and 6V was closely related to 7V. Further analysis was conducted with the available DNA sequences and the deduced amino-acid sequence of each ORF (Table 1 a). DNA analysis showed that the homologies in the five primate bocaviruses were 76·2–91·0% and the homologies in the animal bocaviruses were 33·7–93·6%. With respect to the deduced amino-acid sequence homologies, the primate viruses exhibited 73·3–94·4% for ORF1, 79·8–90·6% for ORF2 and 68·4–87·9% for ORF3 while the animal viruses exhibited 36·7–93·3% for ORF1, 15·3–93·3% for ORF2 and 7·3–88·6% for ORF3. Thus, ORF1 and ORF2 are more conserved than ORF3 in their respective virus clusters.
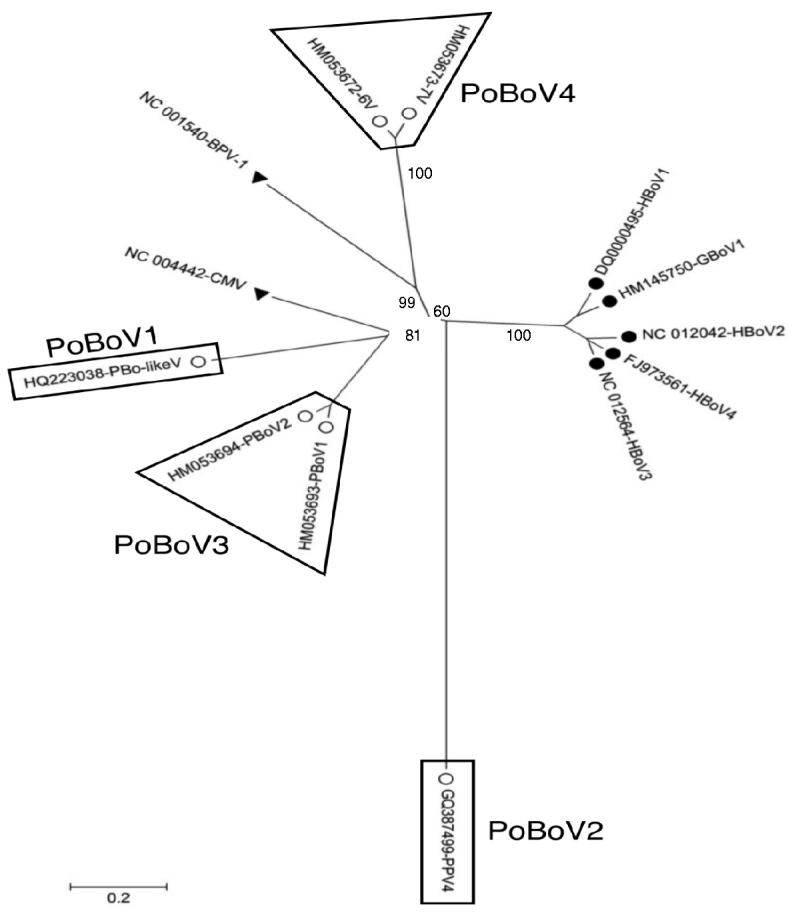
Fig. 1. Phylogenetic distances of bocaviruses based on the deduced amino-acid sequence of ORF2. Primate bocaviruses (•), porcine bocaviruses (○), and CMV and BVP-1 (▾).
Table 1a. Nucleic acid and amino-acid sequence homologies of bocaviruses
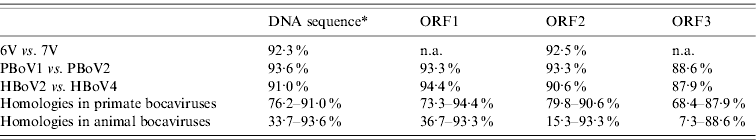
ORF1, ORF2, ORF3, Deduced amino-acid sequence; n.a., not available.
* Almost complete genome DNA sequences except 6V and 7V which only had around 2400 bp.
Recently, a classification method based on the complete ORF2-encoded VP1 was proposed to characterize the human bocaviruses [Reference Kapoor8]. According to this proposal, viruses showing >8% protein and >10% nucleotide difference in the VP1 sequences should be considered different species, while viruses showing >1·5% protein and >5% nucleotide difference should be considered a different genotype. Based on this classification, the human viruses HBoV2 and HBoV4 belong to two different species, while the porcine viruses PBoV1/PBoV2 and 6V/7V belong to two separate species each having two different genotype viruses (Table 1a). Taken together, the six PoBoVs should be classified into four species. We have chosen the PoBoV nomenclature to avoid species/genotype confusion in current nomenclature. The four species are: PoBoV1 (PBo-likeV), PoBoV2 (PPV4), PoBoV3 (PBoV1/PBoV2) and PoBoV4 (6V/7V), with PoBoV3 and PoBoV4 having two genotype viruses each.
A total of 166 clinical samples (64 sera, 102 tissues) were collected from 128 sick pigs, and 38 serum samples were collected from clinically healthy breeding pigs. These samples were collected during January-November 2010 from ten Chinese provinces – Anhui, Fujian, Guangxi, Henan, Hubei, Jiangsu, Jiangxi, Shandong, Shanghai and Zhejiang. PCR primers were designed to detect PoBoVs, porcine reproductive and respiratory syndrome virus (PRRSV), classic swine fever virus (CSFV) and porcine circovirus type 2 (PCV2) individually. The detection methods for PBoLV, PPV4, PRRSV, CSFV and PCV2 have been described in previous studies [Reference Huang13, Reference Zhai15–Reference Zhang18]. Four sets of primers were designed for the detections of PBoV1, PBoV2, 6V and 7V viruses. The primers for PBoV1 (5′-GACCAAGTCCTACGCCATCA-3′/5′- GTTCTCCAATGTCGCCTTTT- 3′) were expected to yield a PCR product of 430 base pairs (bp) at nt 898–1327. The primers for PBoV2 (5′-GCCTCAGTGGCATTATGGAAGAT-3′/5′-TACAGAGCAGGTAGTTGACGATGAA -3′) were expected to yield a PCR product of 428 bp at nt 395–822. The primers for 6V (5′-GCGGGCTTCTGTTTCCTGGTTAT-3′/5′-GAGATGCCGACGCCGTGGTTGTT-3′) were expected to yield a PCR product of 517 bp at nt 357–873. The primers for 7V (5′-CCAGCGAGAACGGAACCAAC-3′/5′-TCCCGTGGCGTAAATGTTGA-3′) were expected to yield a PCR product of 527 bp at nt 1662–2188.
Two hundred microlitres of tissue homogenate or serum was used for viral DNA extraction using a Virus Genome Extract DNA kit according to the manufacturer's instructions (catalogue no. DP315, Tiangen Biotech Inc., Beijing, China). The PCR reaction was performed according to the manufacturer's instructions (catalogue no. KT201-2, Tiangen Biotech Inc.). Briefly, the 25 μl PCR reaction mixture contained 0·5 μl of each forward and reverse primer (10 μm), 12·5 μl of Taq mixture (containing 0·1 U Taq DNA polymerase, 0·4 mm of each dNTP, 2× concentration Taq buffer), 10·5 μl ddH2O and 1 μl sample DNA. The cycling programme for PBoV2, 6V and 7V PCR was: pre-denaturation at 95°C for 5 min, 40 cycles at 94°C for 30 s, 60°C for 30 s, and 72°C for 40 s, with a final extension for 10 min at 72°C. With PBoV1, the annealing temperature was changed to 54°C. In order to ensure that the PCR fragments were derived from the target virus, 2–4 PCR products of each virus were gel-purified and sequenced directly or they were cloned into the PCR2.1T/A vector (lot no. 8210169, Invitrogen Corporation, USA) for sequencing. Nucleotide sequences were determined using an AB-3730 automated DNA sequencer and deposited in GeneBank (accession numbers: HQ910436–HQ910449; organism: bocavirus pig; location: viron; collection date: 2010). The obtained sequences were analysed with the BLASTx program (http://www.ncbi.nlm.nih.gov/ blast).
The results showed that all four PoBoV species were highly prevalent in Chinese swine herds (Table 1 b). The incident rates of PoBoV1-4 were 28·9%, 6·6%, 19·3% and 39·7%, respectively. Thus, PoBoV4 (6V/7V) had the highest rate and PoBoV2 (PPV4) had the lowest rate. The distribution of PoBoVs between healthy/sick animals also varied: PoBoV1 (23·7%/30·5%), PoBoV2 (15·8%/3·9%), PoBoV3 (10·5%/21·9%), and PoBoV4 (44·7%/38·3%) (note: the rates of PoBoV3 and PoBoV4 are not the addition of those rates of PBoV1/2 and 6V/7V; for dual co-infections of PBoV1/2 and 6V/7V the rates need to be subtracted, respectively). Dual co-infections of PoBoVs were quite common (15 combinations) with rates ranging from 1·2% to 16·9%. Furthermore, multiple co-infections were also detected. We detected seven combinations of 3-virus infections: PBoLV/7V/PBoV1 (9/166), PBoLV/7V/PBoV2 (10/166), PBoLV/7V/6V (12/166), PBoLV/6V/PBoV1 (6/166), PBoLV/6V/PBoV2 (10/166), 7V/6V/PBoV1 (8/166), 7V/6V/PBoV2 (10/166); one 4-virus infection: 7V/6V/PBoV1/PBoV2 (4/166) and one 5-virus infection: PBoLV/7V/6V/PBoV1/PBoV2 (3/166). In addition, co-infections of PoBoVs with PRRSV, CSFV and PCV2 were quite common. The presence of PRRSV, CSFV and PCV2 was detected in the 166 samples examined: 33/131 (25·2%), 29/98 (29·6%) and 61/103 (59·2%) samples were PRRSV, CSFV and PCV2 positive, respectively. Of these samples, PoBoVs co-infected with PRRSV, CSFV and PCV2 were, 0–45%, 6·9–34·5% and 3·3–39·3%, respectively.
Table 1b. Porcine bocaviruses: infection rates, dual infections and co-infections with PRRSV, CSFV and PCV2
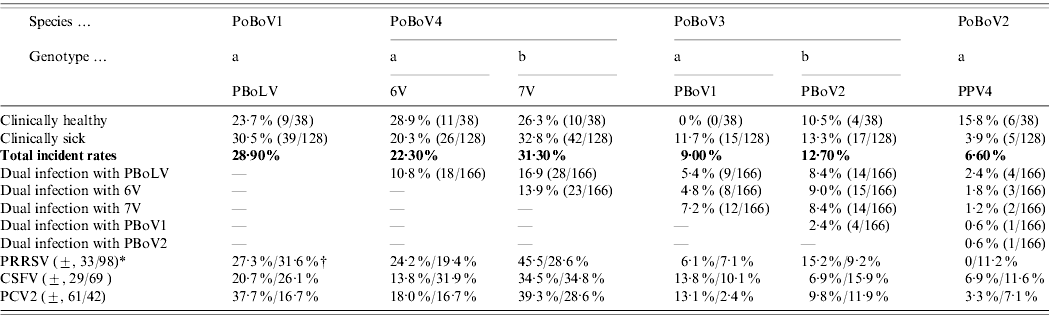
* (±, 33/98)=A total of 131 samples were tested for PRRSV, and 33 were positive, 98 were negative.
† (27·3%/31·6%)=Nine out of 33 (27·3%) PRRSV-positive samples were PBoLV-positive, and 31/98 (31·6%) PRRSV-negative samples were PBoLV-positive.
Considering the dual infection of PBoV1 and PBovV2, the total rate of PoBoV3 is not 9%+12·7%=21·7% but 21·7% – 2·4%=19·3%. Calculation methods for the total rate of PoBoV4 and rates in both healthy and sick animals were the same.
The Bocavirus genus differs from other genera of the Parvovirinae subfamily in which its genome encodes three ORFs instead of two. Currently, bocavirus or boca-like viruses have been identified in five animal hosts: cow, dog, human, gorilla and pig. Of these viruses, porcine bocaviruses exhibit the most genetic diversity. In fact, based on almost full-length DNA sequence analysis, PPV4 was shown to be most related to bovine parvovirus 2 (BPV2), phylogenetically [Reference Cheung12]. BPV2 is not a bocavirus and it contains only two ORFs. However, based on the deduced amino-acid sequence of ORF2 (VP1) classification [Reference Kapoor8], the six reported porcine bocaviruses should be classified into four species (PoBoV1-4).
Our results revealed that PoBoVs were prevalent in Chinese swine. The total incidence rates of the four PoBoV species from sick and healthy animals in the pigs examined were PoBoV1 (28·9%), PoBoV2 (6·6%), PoBoV3 (19·3%) and PoBoV4 (39·7%). In this study, PoBoV1, PoBoV3and PoBoV4 were more prevalent in sick animals than in their healthy counterparts. With respect to PoBoV2, the present study shows that the virus was less prevalent in sick than in healthy animals (3·9% vs. 8%). This is in contrast to our previous finding, where PoBoV2 (PPV4) was more prevalent in sick than healthy animals (2·09% vs. 0·76%) [Reference Huang13]. It is possible that this difference was due to the source of the animals. Whereas the samples of healthy pigs in the present study were obtained from breeding pigs, the samples of sick pigs in both this and our previous study were obtained from commercial pigs, most of which were tissue samples. The data also indicated that PoBoV2 was more prevalent in breeding pigs than commercial pigs, and PoBoV2 was becoming more prevalent in commercial pig farms (3·9% in 2010 vs. 2·09% in 2009).
The data showed that PoBoV co-infections are rampant in the Chinese swine population. Co-infections with two, three, four or five PoBoVs were detected in the sample examined. Moreover, mixed infections with viruses from different virus families were quite common. But the infection patterns were complex. We detected more PoBoV1 in the CSFV or PRRSV negative samples than in the CSFV or PRRSV positive samples. Interestingly, while more PoBoV-1, -3 and -4 were detected in the PCV2-positive samples, more PoBoV2 was found in the PRRSV-, CSFV- and PCV2-negative samples. Whether the distribution of these viruses in clinically sick or healthy animals has any significant meaning is unknown. However, it is quite clear that more work is needed to delineate the disease-causing potential of these newly discovered porcine bocaviruses and the interactions they may have within themselves or with other viruses from a different virus family.
ACKNOWLEDGEMENTS
The authors thank Dr Marcus Kehrli at USDA-NADC for critical review of the manuscript. This project was partially sponsored by a Basic Research Project grant from the National Nonprofit Research Institutions (2010BJ05) to J.X.L., and National Project funding from the Ministry of Science and Technology (MOST, 2006BAD06A01) to S.S.Y.
DECLARATION OF INTEREST
None.





