INTRODUCTION
Pathogenicity in Escherichia coli strains is determined by the presence of specific virulence factors, such as toxins and adhesins, that help overcome host defences and facilitate colonization, resulting in the development of intestinal and extra-intestinal disease [Reference Kaper, Nataro and Mobley1–Reference Albert4]. The significance of toxins and pathogenicity determinants in those E. coli strains responsible for infections at various body sites is being elucidated through molecular and tissue culture studies [Reference Hoffmann and Schmidt5]. Such work should enable commensal and pathogenic strains to be distinguished and therefore assist diagnosis and treatment.
The characterized toxins include the cytotoxic necrotizing factors (CNFs) and cytolethal distending toxins (CDTs), both of which were first isolated from enteric pathogens from humans [Reference Johnson and Loir3, Reference Caprioli6]. Two major classes of CNF toxins have been identified [Reference De Rycke7]. Both cause profound reorganization of the cytoskeleton characterized mainly by the irreversible formation of thick bundles of actin stress fibres which inhibits cell division [Reference Fiorentini8]. CNF1 causes the enlargement, multinucleation and rounding of HeLa cells, and has been demonstrated in haemolysin-producing E. coli strains isolated from humans with septicaemia and enteritis in Italy [Reference Caprioli6, Reference Caprioli9] and from pigs with diarrhoea [Reference Penrith and Henton10]. CNF2 causes enlargement, multinucleation and elongation of HeLa cells, and has been demonstrated largely in E. coli strains isolated from calves and lambs with diarrhoea [Reference De Rycke, Guillot and Boivin11] and septicaemia [Reference Smith12]. CNF1 strains have mostly been isolated from extraintestinal infections from humans with urinary tract infections (UTIs) and occasionally children with diarrhoea. The majority of CNF2 strains have been isolated from calves with septicaemia or diarrhoea [Reference Tavechio13].
CDT-producing E. coli strains were first described by Johnson & Lior [Reference Johnson and Loir3] who isolated them from children with enteritis. Currently four CDTs have been differentiated by gene sequence. The CDTs, or close homologues, are also produced by other bacteria such as Campylobacter spp. [Reference Pickett14], Shigella dysenteriae [Reference Okuda15] and Haemophilus ducreyi [Reference Cope16]. When tested in HeLa cells, CDTs produce giant mononucleated cells caused by an irreversible block in the cell cycle at the G2/M stage [Reference Comayras17]. Cellular death follows this elongation. As with CNFs, there is debate as to whether CDT-harbouring E. coli are pathogenic to humans. For example, in a case-control study of CDT-producing E. coli in Bangladeshi children Albert et al. [Reference Albert4] found that, although CDT-positive E. coli were isolated from more children with diarrhoea than healthy controls, the difference was not statistically significant.
Additional toxins, haemolysins (hly), commonly present in pathogenic E. coli strains, are also thought to contribute to the virulence of these strains [Reference Caprioli9, Reference Blanco18]. A number of adhesins have been described in pathogenic E. coli strains associated with intestinal and extraintestinal diseases. Adhesins are either associated with fimbrial cell surface structures such as P-fimbriae, S-fimbriae and F17, or not associated with fimbriae and designated as afimbrial adhesins (afa). These factors are encoded by the pap, sfa, F17 and afa related gene clusters respectively [Reference Bertin19–Reference Mazouari22]. Furthermore, the CNF toxins have been thought to be elaborated only by E. coli strains. However, it has been reported that cell extracts from Yersinia pseudotuberculosis induced multinucleation in Hep-2 cells in a manner similar to the effect of CNF caused by E. coli strains. The nucleotide sequence of the Yersinia CNF gene was found to be 65% similar to the CNF gene of E. coli strains [Reference Lockman23].
This study investigated the incidence of CNF- and CDT-harbouring E. coli strains isolated from patients and controls in Northern Ireland using PCR assays for the specific detection of CNF and CDT toxins and other pathogenicity factors. The results will enable the design of studies of suitable power to assess the importance of CNF and CDT production to clinical condition and outcome, since their relevance currently remains unclear.
MATERIALS AND METHODS
Bacterial strains
E. coli isolates from a total of 544 humans were used in this study. Isolates were cultured from the blood (n=266) and faecal (n=237) samples of adult patients in Belfast City Hospital, a 900-bed teaching hospital. Faecal samples were examined diagnostically for a variety of pathogens by standard methods, but only E. coli isolates from them are reported in this study. For the purposes of this study blood isolates were treated as individual samples and not collated into a series for individual patients to exclude incidental contamination, although this is the hospital practice for case diagnosis of septicaemia. Blood cultures were not done for patients with diarrhoea unless septicaemia was also suspected, and it would have been unethical to perform venepuncture in the absence of this suspicion. No controls for blood isolates were available since the blood of healthy individuals is sterile. More than 80% of the faecal samples originated from adult hospital patients, the remainder being submitted by local general practitioners and included children's specimens. To maintain confidentiality, patient records were not accessed. As a consequence of this, it is possible that a few samples were repeat cultures from the same patient. The number of repeat patient samples in this hospital is <1%. The case mix of the hospital is such that around 10–20% of the blood samples came from immunocompromised haematology, oncology and renal patients where it is recognized that it is common for septicaemia to originate from intestinal microflora. Paired specimens of blood and urine would not have been taken in all cases we examined, and confidentiality precludes this being investigated retrospectively. E. coli isolates were processed on arrival at the laboratory, or after overnight storage at 4°C. Cultures from fresh patient specimens (peri-anal swabs for asymptomatic control subjects, age range 19–60 years, who worked in food and veterinary microbiology laboratories, n=41) were subcultured overnight on blood agar plates at 37°C. Control volunteers were not phlebotomized for ethical reasons. Single colonies were tested by multiplex PCR as described previously [Reference Kadhum24] and HeLa cell cytotoxicity assays. Positive strains were confirmed as E. coli by biochemical tests with the API 20E system (bioMérieux, Basingstoke, UK).
Cytotoxicity assay
E. coli strains were grown on blood agar overnight at 37°C. One loopful of colony growth was suspended in 0·5 ml sterile phosphate buffered saline (PBS; 0·1 m, pH 7·2). The suspension was frozen at −70°C and thawed at room temperature, three times. After centrifugation at 10 000 g for 10 min, the supernatants were stored at −20°C until used. The cytotoxicity assay was performed with HeLa cells as previously described [Reference Johnson and Loir3].
Haemolysin PCR assay
E. coli strains were cultured overnight at 37°C on 5% washed bovine blood agar, and haemolytic areas larger than the overlying colony were recorded as positive. Positive haemolysins were tested for α-haemolysin by using specific primers [Reference Fernandez-Prada25]. Amplifications were performed in a 25 μl reaction mixture containing 2–2·5 mmol/l MgCl2, 0·32 mmol/l of each dNTP, 1·2 μmol/l of the primer pair and 0·5 U Taq DNA polymerase in PCR buffer (10 mmol/l Tris–HCl, 50 mmol/l KCl; pH 8·3). The thermal reaction parameters for the PCR were 94°C for 5 min for 1 cycle, followed by 30 cycles of denaturing at 94°C for 1 min, annealing at 48°C for 1 min and extension at 72°C for 1 min before a final extension step at 72° C for 10 min.
PCR assays for other pathogenicity determinant genes
Primed sets are listed in Table 1. Amplifications were performed under the conditions used above except that annealing was at 63°C for 1 min.
Table 1. Primer sequences used for PCR assays
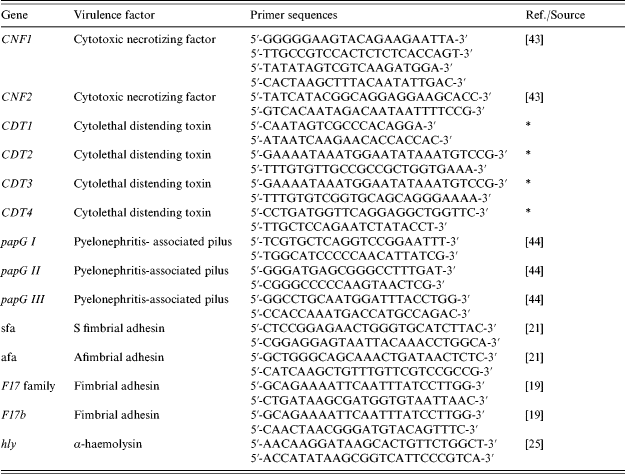
* Gift from Eric Oswald.
Serogrouping
The somatic (O) antigen of E. coli strains was determined by slide agglutination using a collection of 71 antisera that were available at the Veterinary Sciences Division where this work was carried out. Hospital isolates are not routinely serotyped. This collection was established largely for the identification of strains of veterinary significance and consisted of O1, O2, O3, O4, O5, O6, O7, O8, O9, O10, O11, O15, O17, O18, O20, O21, O22, O26, O32, O35, O39, O43, O45, O54, O55, O64, O65, O71, O73, O75, O78, O81, O82, O86, O87, O88, O91, O92, O100, O101, O103, O105, O108, O109, O111, O113, O114, O115, O116, O117, O118, O119, O121, O123, O125, O126, O128, O130, O132, O137, O138, O139, O141, O145, O146, O147, O149, O153, O157, O162 and O168.
RESULTS
Table 2 shows a total of 74/266 (27·8%) blood strains were PCR positive for a toxin [95% confidence interval (CI) 22·5–33·6]. Fifty-two of these (20%) had CNF only, 18 (7%) had CDT only, and four (2%) had both toxin groups. From the 237 faecal strains, 17 (7%, 95% CI 4–11) were toxin PCR positive, of which nine (4%) had CNF only, four (2%) had CDT only and 4 (2%) had both toxin groups.
Table 2. Cytotoxic necrotizing factor (CNF) and cytolethal distending toxin (CDT) producing E. coli strains from human blood and faecal samples
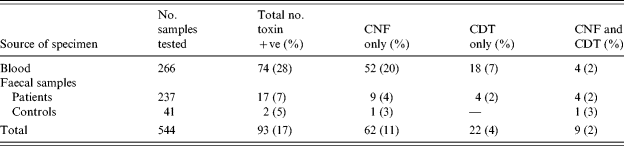
Table 3 summarizes the results obtained with the CNF-positive strains, using the multiplex PCR for the specific detection of CNF1 and CNF2 and CDT1-4. All 52 CNF-producing blood strains were CNF1, three were CNF1/CDT1 and one was CNF1/CDT4. Eighteen of the blood strains were CDT only, 16 with CDT1 and two with CDT4. Of the 13 CNF encoding faecal strains, nine were CNF1, two were CNF1/CDT1 and two were CNF2/CDT3. Another four faecal strains were CDT4 only. Two toxin-positive strains were isolated from the control faecal samples. One was CNF1 and the other was CNF2/CDT3. All of the PCR toxin results were confirmed by the HeLa cell cytotoxicity assay, for both CNF and CDT toxins.
Table 3. Distribution of papG, sfa, afa, F17 and hly in CNF1, CNF2, CDT1-4 positive E. coli strains
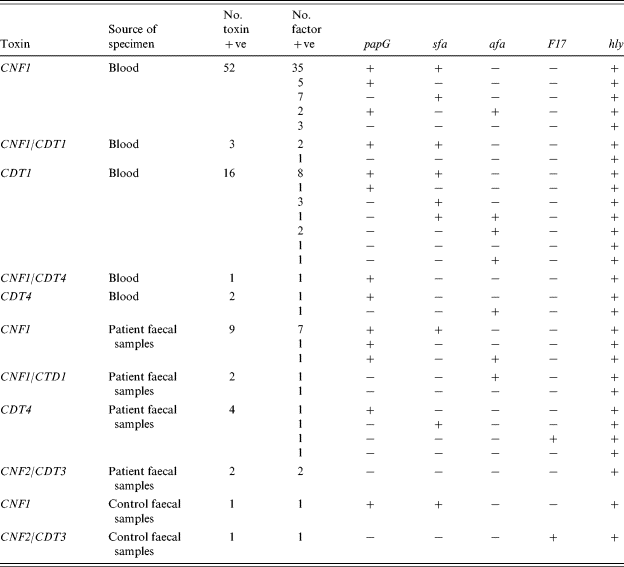
Genes for toxins and virulence factors were much more common in patient blood specimens than control faecal samples. Only two of the 41 healthy control faecal samples (4·9%, 95% CI 0·6–16·5) possessed these toxin genes. The relative risk of having either CNF or CDT toxin genes in patient compared to control faecal samples was 1·47 (95% CI 0·35–6·13; Fisher's exact test). Although there is no statistical evidence of an increased risk (P=0·45), the 95% confidence interval is very wide and includes the possibility of a true sixfold increased risk. The relative risk of having either CNF or CDT toxin genes in blood compared to faecal isolates was 3·88 (95% CI 2·36–6·38). This was highly significant (P<0·0001) and reflects the greater importance of E. coli isolates as a cause of bloodstream infections than in faeces where they may be less likely to be a cause of symptoms. The limited number of control samples gave little power to detect differences between patients and controls, and a larger study would be informative.
The presence of genes encoding F17 fimbriae, papG, sfa, afa and the expression of α-hly in the CNF1-, CNF2-, and CDT-producing strains is also summarized in Table 3. Most factor patterns contained five or fewer strains. Three characteristic factor patterns containing more than five strains were prominent. A group of 35 were from the CNF1 patient blood strains, seven from patient faecal samples, and one from control faecal samples (CNF1, papG, sfa, hly). A group of eight were from the CDT blood strains (CDT1, papG, sfa, hly), and a group of seven were from the CNF1 blood strains (CNF1, sfa, hly).
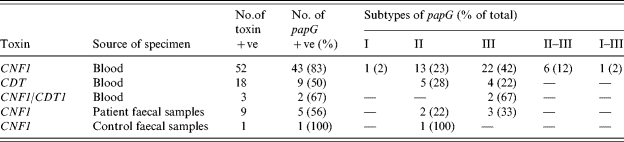
Table 4 summarizes the subtypes of the papG gene detected in the toxigenic strains. Their distribution in the toxigenic strains, in descending order, was III, II and both II and III. In addition one strain possessed I, and one strain had I and III. These subtypes were randomly distributed within the papG-positive strain divisions listed in Table 3.
Table 4. The subtypes of papG expressed by CNF1- and CDT-positive E. coli strains
Serotypes O2 (9), O4 (3), O6 (17), O18 (1), O22 (2), O26 (2), O27 (3) and 19 untypable were detected in the CNF1 blood strains. Eleven (31%) of the O6 strains were from the large 35 encoding pattern group of strains described above. Serotypes O2 (7), O18 (3), O6 (1) and seven untypable were detected in the CDT blood strains. Four of the O2 strains and two of the O18 serogroups were from the similar encoding pattern group of 16 strains described above. The CNF2 strains were O75 and O147.
Data concerning the age of the septicaemic cases were available. Analysis showed that the majority of the cases were in adults aged >50 years. From 52 CNF1 and CNF1/CDT1 and/or CNF1/CDT4 cases and 18 CDT cases, 49 were in this age group, respectively. Similarly, from 192 cases from which toxigenic strains were not isolated, 140 involved adults aged >50 years. Insufficient information was available to make a meaningful analysis of diarrhoea cases by age.
DISCUSSION
Common and specific PCR primers were employed to amplify genes encoding the CNF and CDT toxins, α-hly and adhesins associated with these toxins. The PCR methods were specific for the detection of E. coli strains encoding these virulence factors. Although the factors examined have been previously implicated as virulence determinants, to the best of our knowledge, this is the first investigation of the presence of all of them in large numbers of strains isolated from clinical specimens. Several patterns of factors were detected in the strains examined, but the presence of common characteristic patterns in 35 (67%) of the CNF1 blood strains and eight (50%) of the CDT blood strains are indicative of strain types with greater invasive potential. Additional toxigenic strains were apparent in the faecal samples, and the significance of the dominant factor pattern observed with seven (64%) of the CNF1 strains was also persuasive. CNF1 and CNF2 are 85·7% genetically identical [Reference Oswald26]. The CNF1 and CNF2 specific primers used in this study amplified unique sequences of different molecular size, enabling them to be used in a multiplex PCR. This assay enabled the differential detection of each toxin in the strains tested. Strains expressing both toxins have never been recorded. In the present study, two CNF2 strains belonging to serogroup O75 and O147 were recorded. CNF2 has largely been detected in strains isolated from polygastric animals, and the predominance of CNF1 strains from human sources in the present study supports the results of other studies [Reference Caprioli6, Reference Bisicchia27, Reference Blanco28].
The association of the CNF1-producing strains in this study with the other factors confirms the work of others [Reference Caprioli6, Reference Blanco28–Reference Germani32]. The combination of CNF1, α-hly and P-fimbriae genes has been demonstrated in the human uropathogenic strain, J96, possibly as a pathogenicity island [Reference Blum33]. These factors, together with sfa, were the predominant combination observed in 35 (67%) of the toxigenic blood strains. In addition, α-hly, P-fimbriae and sfa were also the factors encoded in the predominant CDT-expressing blood strains, suggesting a role in invasion for these toxin-associated factors. Seven CNF faecal strains also encoded the same pattern of associated factors. UTIs and septicaemia are frequently caused by gastrointestinal microorganisms and binding of P-fimbriae to the Galα1→4Galβ disaccharide confers binding not only to urinary tract epithelial cells but also colonic epithelium. This is consistent with UTIs originating from faecal contamination.
The F17b fimbriae are well documented as an additional factor encoded by genes present in the CNF2 Vir plasmid [Reference Oswald34]. The absence of F17b fimbriae in the strains of this study was as expected. Although one blood CDT strain in this study was positive for the F17 family, the absence of F17b in CNF1 strains was confirmed.
The significance of CNF1 and α-hly has been evaluated in animal experiments employing genetically defined and mutant strains [Reference Elliott35, Reference Wray36]. The results suggested that these virulence factors induced pathological changes, including intestinal inflammation, giving rise to enterocolitis and bloody diarrhoea. Although outbreaks and cases of diarrhoea associated with CNF1 strains have been reported [Reference Caprioli6, Reference Blanco37–Reference Necoletti39], the low incidence of CNF1 faecal strains in the present study indicates its limited importance in the population investigated, and probably more widely.
A low incidence of CNF-encoding strains has been detected in faecal samples collected from healthy humans [Reference Bisicchia27, Reference Blanco28]. Similar results were obtained in a preliminary study at this laboratory, where one CNF1 and one CNF2/CDT3 strains were isolated from 41 samples. Whether such isolates are potentially pathogenic strains in asymptomatic carriers or are non-pathogenic strains cannot be determined with the information currently available. In general, factors that are associated with virulence are more commonly detected in strains isolated from patients than from healthy humans.
Various studies have reported the isolation of CDT-expressing strains from patients with a variety of diarrhoea symptoms and encephalopathy [Reference Albert4, Reference Anderson and Johnson40]. Evidence from the use of a suckling mouse model suggests that the CDT toxin group causes secretory diarrhoea and necrosis of the colonic epithelium [Reference Okuda15]. It is not clear whether these effects occur in human disease, and one study has suggested that CDT-producing E. coli strains are not associated with diarrhoea, as no statistical difference was found between their isolation from children with or without diarrhoea [Reference Albert4]. In the present study, there was a low prevalence of CDT strains isolated from faecal samples, 4/236 (2%), indicating their relative unimportance in Northern Ireland.
The association of CDT strains with papG, sfa, F17b and α-hly was highly variable in this study. The commonest pattern of factors displayed by eight of the 16 blood strains, was papG, sfa and α-hly positive. The pathogenic significance of this combination of factors remains to be clarified.
The serotypes found in the toxigenic strains in this investigation have been recorded in other studies which have reported that CNF1 strains belong to a limited number of serogroups, and that type O6 was the most common [Reference Caprioli6, Reference Giugliano41]. In this investigation O6 strains were detected most commonly among CNF1 strains, and O2 and O18 strains were found to be the most frequently identified in CDT strains.
The role of the CNF and CDT E. coli strains in septicaemia and/or diarrhoea in humans is not fully understood. A significant proportion of blood isolates may have originated from UTIs, particularly in elderly female patients where around half of bacteraemias are UTI related. The highest incidence of toxic blood strains was among elderly patients (aged >50 years). In this group, they are probably opportunistic pathogens. The finding of E. coli in blood is of clinical significance, but a small proportion of isolates may have been skin contaminants. Foodborne transmission may possibly occur [Reference Smith42], leading to a transient or colonizing alteration of the gastrointestinal flora. With accessory factors, these bacteria may give rise to UTIs, bacteraemia and septicaemia in a small proportion of cases. This work identifies the importance of these E. coli strains in bloodstream infections and reports the frequencies of genes occurring in E. coli from patients' blood and faecal specimens. It provides baseline values that will assist the development of studies targeted more specifically to establish the clinical relevance of these pathogenicity determinants.
ACKNOWLEDGEMENTS
The provision of the E. coli clinical isolates by Barry McClurg, and critical comments by Dr Paul Rooney, Belfast City Hospital, Belfast, are gratefully acknowledged. The assistance of John Early, Food Microbiology Division, Newforge Lane, Belfast, and staff members of the Molecular Bacteriology and the Salmonella Laboratories at the Veterinary Sciences Division are also recognized. The authors are grateful for the supply of CDT1, 2, 3 and 4 primer sequences by Dr Eric Oswald (Laboratoire Associé INFRA-ENVT de Microbiologie Ecole Nationale Vétérinaire, Toulouse, France), and Nick Andrews (Health Protection Agency) for statistical advice.
DECLARATION OF INTEREST
None.






