Escherichia coli isolates from humans and livestock are increasingly resistant to third- and fourth-generation cephalosporins, such as cefotaxime, ceftazidime and cefepime. The major cause of this resistance is the expression of plasmid or chromosomal located genes encoding for either extended-spectrum-β-lactamases (ESBLs) or for AmpC-β-lactamases [Reference Carattoli1–Reference Woodford, Turton and Livermore3]. The frequency of individual ESBL genotypes and their association with certain E. coli lineages and clones is undergoing constant change. Over the past 20 years, ESBLs of the genotype CTX-M have emerged as predominant ESBLs in E. coli worldwide [Reference Mathers, Peirano and Pitout4, Reference D'Andrea5]. From the >100 different CTX-M variants described so far, CTX-M-15 is currently the most frequent genotype identified from ESBL-producing E. coli [Reference Nicolas-Chanoine, Bertrand and Madec6, Reference Banerjee and Johnson7]. CTX-M-15 is often associated with sequence type (ST) 131 and serotype O25b, which is the predominant lineage of extraintestinal pathogenic E. coli in recent years [Reference Mathers, Peirano and Pitout8, Reference Rogers, Sidjabat and Paterson9]. In a recent Germany-wide study, the ESBL genotypes and multilocus sequence types of 233 ESBL-producing E. coli isolates obtained from German hospitals and medical practices in 2011–2012 were determined [Reference Pietsch10]. Next to CTX-M-15 (50·4%), the genotypes CTX-M-1 (28·4%) and CTX-M-14 (5·6%) were found to be the most common variants. In this study we characterized ESBL-producing E. coli isolates from a single centre, the University Medical Center Göttingen (UMG) in south Lower Saxony, Germany. The UMG is a maximum-care hospital with a capacity of about 1500 beds. Annually around 60 000 patients are admitted and about 173 000 patients are seen in the different outpatient departments.
During November 2013 and May 2014 all E. coli isolates from UMG with a cefotaxime resistance >2 mg/l and/or a ceftazidime resistance >4 mg/l were collected. Species identification was performed by MALDI-TOF mass spectroscopy (Bruker, Germany). Antibiotic resistance was determined by VITEK 2 analysis (bioMérieux, France). The collected isolates were adjusted for re-isolates from the same patient, resulting in a total of 160 isolates (Table 1), 79 (49·4%) of which were isolated from urine. The median age of patients was 72 years. Multilocus sequence typing analysis was performed as previously described by amplification and sequencing of fragments from seven housekeeping genes [Reference Wirth11]. The sequence type was determined using the University of Warwick database (http://mlst.warwick.ac.uk/mlst/dbs/Ecoli). ST131 occurred with an overall frequency of 24% (n = 39) in the collection. Table 1 shows the ST131 proportion for the individual clinical specimens. The specimens with the highest ST131 prevalence were rectal swabs (42·9%, n = 14), groin swabs (37·5%, n = 8) and urine (29·1%, n = 79). The remaining 76% (n = 121) belonged to 51 different sequence types. The most prevalent non-ST131 were ST101 (5·0%, n = 8), ST58 (5·0%, n = 8), ST10 (4·4%, n = 7), ST38 (4·4%, n = 7), ST410 (3·8%, n = 6) and ST453 (3·1%, n = 5) (Table 2). A list of all identified sequence types is provided in Supplementary Table S1.
Table 1. Clinical specimens of the isolates (n = 160)*

* Median age of all patients: 72 years.
† Median age of patients with ST131 isolates: 71 years.
‡ Nine of 39 ST131 isolates were screening isolates.
§ Median age of patients with non-ST131 isolates: 72 years.
|| Twenty of 121 non-ST131 isolates were screening isolates.
Table 2. CTX-M-type distribution for the most abundant sequence types (n > 3) from ESBL-producing E. coli

Values given are percentages.
* In addition to the shown CTX-M types, one ST38 isolate was positive for CTX-M-55 and one ST410 isolate was positive for CTX-M-17. Two ST73 isolates were CTX-M negative.
At least four (ST10, ST38, ST58, ST410) out of these six major sequence types have been previously described to be associated with animals and livestock farming [Reference Pietsch10, Reference Fischer12–Reference Schaufler15]. Lower Saxony is known as important livestock producing state within Germany [Reference Cuny, Kock and Witte16]. ST38 was also reported to be isolated frequently from healthy humans [Reference Valenza17]. Interestingly, the two most abundant non-ST131 sequence types (ST101 and ST58) were identified only at a very low frequency in a Germany-wide surveillance study [Reference Pietsch10], indicating that significant local differences can occur in sequence-type distribution of ESBL-positive E. coli. Outside Germany, CTX-M-positive ST58 were isolated from various animals, e.g. dogs, rooks and poultry [Reference Damborg18]. The globally successful ST101 was found to be associated with the metallo-β-lactamase NDM-1, which confers carbapenem resistance, but was also shown to possess less virulence factors than ST131 [Reference Peirano19]. ST453 was described as an emerging sequence type associated with extraintestinal infections, particularly urinary tract infections throughout the world [Reference Goldstone20].
CTX-M genotyping was performed according to Strauss et al. [Reference Strauss21]. In short, a bla CTX-M multiplex polymerase chain reaction (PCR) with four primer pairs for bla CTX-M-1, bla CTX-M-2, bla CTX-M-9, and bla CTX-M-8/25 was performed (Supplementary Table S2). PCR products were analysed on a 1·5% agarose gel and purified using the Qiagen PCR purification kit (Qiagen, Germany). After DNA sequencing of the PCR product from both sites (Seqlab, Göttingen), the CTX-M genotype was determined by BLAST-N analysis in the NCBI database. All ST131 isolates (n = 39) and 94·3% (n = 114) of the non-ST131 isolates were CTX-M positive. The predominant CTX-M variants were CTX-M-1 (44·4%, n = 71), CTX-M-15 (34·4%, n = 55), CTX-M-14 (9·4%, n = 15) and CTX-M-27 (5·0%, n = 8) (Fig. 1).
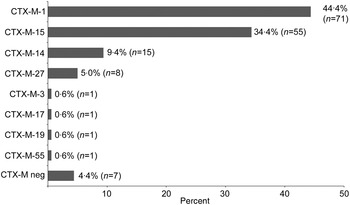
Fig. 1. CTX-M-type distribution in ESBL-producing E. coli. Of 160 isolates 153 (95·6%) were CTX-M positive. Eight different CTX-M variants were identified.
A high prevalence of CTX-M-1, CTX-M-14 and CTX-M-15 is often observed in E. coli with the ESBL phenotype and CTX-M-15 is currently the most frequent CTX-M gene in German healthcare settings [Reference Pfeifer, Cullik and Witte2, Reference Pietsch10, Reference van Hoek22]. The relatively low CTX-M-15 rate of 34·4% in our study appears to be due to the lower ST131 proportion in our collection (24%), compared to a ST131 rate of 35·8% in the Germany-wide study of Pietsch et al. [Reference Pietsch10]. The widely disseminated E. coli clone O25b:H4-ST131 frequently carries the CTX-M-15 gene, while non-ST131 sequence types show, on average, lower CTX-M-15 associations [Reference Mathers, Peirano and Pitout8, Reference Rogers, Sidjabat and Paterson9]. However, the proportion of CTX-M-1, CTX-M-15 and CTX-M-14 in our study is similar to the results of a case-control study performed at the Charité University Hospital in Berlin, Germany, with patients colonized with community-acquired ESBL-positive E. coli [Reference Leistner23].
The individual E. coli sequence types in our study displayed significant differences in the distribution of the various CTX-M types (Table 2). The predominant CTX-M variant in ST131 isolates was CTX-M-15 (56·4%, n = 22), followed by CTX-M-27 (20·5%, n = 8), CTX-M-14 (12·8%, n = 5) and CTX-M-1 (10·3%, n = 4). In contrast, CTX-M-1 was the predominant genotype in non-ST131 isolates, namely in the abundant sequence types ST101 (8/8 isolates), ST58 (5/8 isolates), ST10 (5/7 isolates) and ST453 (5/5 isolates). An association of ST101, ST453 and ST10 with CTX-M-1 was also reported in a recent Germany-wide study [Reference Pietsch10]. From the six most abundant sequence types only ST410 shows an association with CTX-M-15 (5/6 isolates). This association of ST410 and CTX-M-15 was also observed in ESBL-positive E. coli collected from German and Brazilian hospital patients [Reference Pietsch10, Reference Peirano24]. Recent data revealed genetic similarities between human and animal CTX-M-15-positive ST410 isolates and suggest a clonal dissemination of specific E. coli ST410 clades [Reference Falgenhauer25]. The second most common CTX-M variant in our ST131 isolates, i.e. CTX-M-27, was absent in all non-ST131 isolates and was also found at a significantly lower rate (1/127 isolates) in nosocomial patients from other German hospitals [Reference Pietsch10]. CTX-M-27 differs from CTX-M-14 by a single Asp240Gly substitution that was shown to confer higher ceftazidime resistance (minimum inhibitory concentration: 8 vs. 1 mg/l) [Reference Bonnet26]. CTX-M-27 is the predominant allele in ST131 isolates from Japanese hospitals. A recent study reported a CTX-M-27 frequency of 45% in ST131 isolates collected at 10 Japanese acute-care centres [Reference Matsumura27].
In conclusion, our single-centre study in Lower Saxony, Germany reveals a distinct sequence-type distribution of ESBL-producing E. coli compared to the average nationwide sequence-type distribution. This leads as a consequence to a shift in the distribution of CTX-M alleles, with CTX-M-1, not CTX-M-15, as the most frequent allele. Local differences in sequence-type frequency might reflect potential area-dependent differences in the proportion of livestock-associated sequence types, e.g. ST10, ST38 and ST410. The greater abundance of these livestock-associated sequence types found in our study once more reveals the necessity of studying the role of transmission from animals to humans or vice versa.
SUPPLEMENTARY MATERIAL
For supplementary material accompanying this paper visit http://dx.doi.org/10.1017/S0950268816001412.
DECLARATION OF INTEREST
None.






