INTRODUCTION
Leptospirosis is one of the most widespread zoonoses and is caused by infection with pathogenic spirochaetes of the genus Leptospira [Reference Levett1]. Its incidence in humans is most frequent in developing countries, and the spectrum of human disease ranges from subclinical infection to severe symptoms of multi-organ dysfunction with high case-fatality rates, reaching mortalities as high as 70% in the case of severe pulmonary haemorrhage syndrome [Reference Vijayachari, Sugunan and Shriram2–Reference McBride5]. However, its prevalence is still underestimated due to low awareness in the medical community and an absence of specific symptoms and readily available tests. In addition, some patients may also experience transient or mild manifestations [Reference McBride5].
Rodents are recognized as important mammalian reservoirs of Leptospira spp. [Reference Guerra6, Reference Meerburg, Singleton and Kijlstra7]. Infection may occur early in the lifespan of the animal and the chance of infection increases with age [Reference Levett1, Reference Koizumi8]. Rodents only present mild chronic disease or are asymptomatic, and shed infectious organisms in the urine during their lifetime [Reference Koizumi8–Reference Plank and Dean10]. Humans may be infected indirectly by contact with contaminated water, soil or mud in a moist environment, or by direct contact with urine, fresh carcasses or organs [Reference Faine11, Reference Mayer-Scholl12]. Therefore, surveillance on the carriage status of reservoir hosts and analysis of the characteristic of causative agents contribute to the clinical laboratory diagnosis, active surveillance, outbreak investigation and source tracking for leptospirosis.
Human leptospirosis, including death, has been reported in Qiandongnan prefecture, southeast Guizhou, every year in recent time. For example, 127 cases including 28 deaths were reported in Liping county from 2001 to 2008 [Reference Yang13]. However, L. interrogans was not isolated from patients and the aetiological characteristics such as epidemic serogroup and serovar remained unclear.
Pathogenic Leptospira are classified into more than 200 serovars based on serological methods [Reference Levett14, Reference Morey15]. Currently, molecular methods like genotyping by multiple locus variable-number tandem repeat (VNTR) analysis (MLVA) are used for typing leptospires [Reference Majed16–Reference Pavan20]. Zhang et al. [Reference Zhang21] showed that MLVA based on seven loci was suitable for the typing of epidemic reference strains belonging to the ten serogroup of Leptospira in China. In order to track the source of infection and understand the aetiological characteristic of leptospirosis, in 2011 we performed surveillance of field mice carriage in Jingping and Liping counties, an epidemic region located in Qiandongnan prefecture of Guizhou province, China. G1/G2-based PCR was used to confirm the leptospiral isolates. MLVA was applied to type and reveal the genetic diversity and relationships between regional strains. This was intended to aid clinical laboratory diagnosis, active surveillance, outbreak investigation and source tracking for leptospirosis.
MATERIAL AND METHODS
Field mice trapping and leptospire isolation
Field mice were trapped, during September–October, using the Trap-night method [Reference Yalin22] (traps with peanut bait) in rice-field environments in Jinping and Liping counties, Qiandongnan prefecture, southeast of Guizhou province, an area where leptospirosis patients have been reported in recent years. Traps were placed in the evening and collected the next morning. Trapped field mice were identified by genus, species, and gender based on phenotypic characteristics (ears, body, tail, fur colour, sex) [Reference de Faria23]. Field mice were dissected to collect the kidneys according to CERoPath protocols (www.ceropath.org) [Reference de Faria23], which follow animal care health security for field parasitologists and quality data-handling principles. Animal care and manipulation followed the international regulation (American Veterinary Medical Association Council on Research). Live animals were killed by decapitation under anaesthesia by diethyl ether. Freshly isolated kidney samples were inoculated into 8 ml liquid Ellinghausen-McCullough-Johnson-Harris (EMJH) medium (Difco Laboratories, USA). Cultures were incubated at 28°C and evaluated weekly by dark-field microscopy for up to 2 months [Reference Subharat24].
Leptospiral strains and cultivation
Fifteen Chinese reference strains belonging to 15 serovars in 15 serogroups of pathogenic Leptospira spp. were provided by the Chinese Centre for Disease Prevention and Control. The reference strains and isolates were cultivated at 28°C in EMJH [Reference Ivanova25].
Microscopic agglutination test (MAT)
The MAT was performed to detect antibodies in the serum samples against the various Leptospira [Reference Faine11]. A total of nine serum samples from patients from the epidemic area were tested by MAT using the 15 reference strains and the leptospiral isolates. The MAT titre was expressed as the reciprocal of the highest serum dilution that resulted in 50% agglutination of leptospires. Samples with titres ⩾100 were recognized as positive.
DNA extraction
All leptospiral strains used in this study were cultured in EMJH and the bacteria harvested as described previously [Reference Awad-Masalmeh26]. DNA was extracted from cultures of the isolated strains using a DNA extraction kit (SBS Genetech, China) according to the manufacturer's instructions.
PCR confirmation
PCR was used to confirm the four isolated strains using G1/G2 primers for identification of pathogenic Leptospira [Reference Gravekamp27] using a PCR kit (TaKaRa, Japan). Briefly, each 25 μl reaction system contained 2 μl DNA, 12·5 μl PreMix Taq, 2 μl G1 (5′-CTG AAT CGC TGT ATA AAA GT) and G2 (5′-GGA AAA CAA ATG GTC GGA AG) primers at 10 pmol/μl, and 6·5 μl deionized water. Amplification was performed on a Biometra TProfession thermocycler (Germany) at 95°C for 5 min, followed by 35 cycles at 94°C for 60 s, 55°C for 60 s, and 72°C for 90 s, with a final single extension of 72°C for 10 min, and then held at 4°C. Amplified products were characterized by electrophoresis of 1 μl of each reaction on a 1·2% agarose gel for 30 min at 100 V.
MLVA analysis
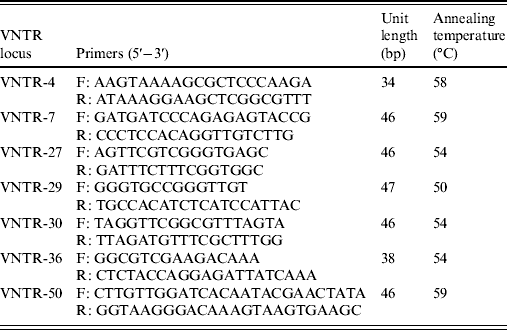
The variable number tandem repeat (VNTR) loci used in this study were the seven loci (VNTR-4, VNTR-7, VNTR-27, VNTR-29, VNTR-30, VNTR-36, VNTR-50) which have been shown to be suitable for the typing of reference strains belong to the ten serogroups of Leptospira in China [Reference Zhang21]. The primers were synthesized by Invitrogen (China). PCR amplification was performed in a final volume of 50 μl. All reactions contained 25 μl PCR Pre Mix (TaKaRa), 2 μl of forward and reverse primers of VNTR locus (10 pmol/μl), 2 μl DNA (1 ng/μl), and 19 μl deionized water. The PCR reactions were run on the Biometra thermocycler using the following conditions: 94°C for 10 min, varying cycles of 94°C for 30 s, annealing at varying temperatures (Table 1) for 30 s, extension at 72°C for 1 min, with a final single extension at 72°C for 10 min, and then held at 4°C. Amplified products were detected by electrophoresis of 1 μl of each reaction on a 2% agarose gel for 2 h at 100 V. Each VNTR locus was identified by the size assigned by Genescan software (Applied Biosystems, USA). The copy number of the repeats of each VNTR locus was deduced from the band sizes of the amplified products. This size was then converted into an allele designation, which in turn formed the allele string for the seven loci. The allele string was constructed in the following order: VNTR4-VNTR7-VNTR27-VNTR29-VNTR30-VNTR36-VNTR50. The Bionumerics software package, version 4.0 (Applied Maths, Belgium) was used for cluster analysis of the data using the categorical coefficient and the unweighted pair-group method using average linkage clustering parameters. The reference strains used for clustering were the ten strains of the Chinese epidemic serovar, which were successfully analysed by Zhang et al. [Reference Zhang21]. The polymorphism indices of individual and grouped VNTRs were calculated using Nei's diversity index.
Table 1. VNTR loci proposed for MLVA of leptospiral isolates

VNTR, Variable-number tandem repeat; MLVA, multiple locus variable-number tandem repeat analysis.
RESULTS
Trapped field mice and isolated leptospires
A total of 118 field mice were trapped, and the most prevalent species in Jinping and Liping was Apodemus agrarius (37·8% of total mice for Jinping and 21·9% for Liping). Three strains of leptospires (JP13, JP15, JP19) were isolated from A. agrarius in Jinping county, and one strain (LP62) from A. agrarius in Liping county (Table 2).
Table 2. Field mice trapped and leptospires isolated at surveillance sites, Jingping and Liping counties, 2011

* Number of leptospires isolated from Apodemus agrarius.
Anti-Leptospira antibodies from patients in the epidemic area
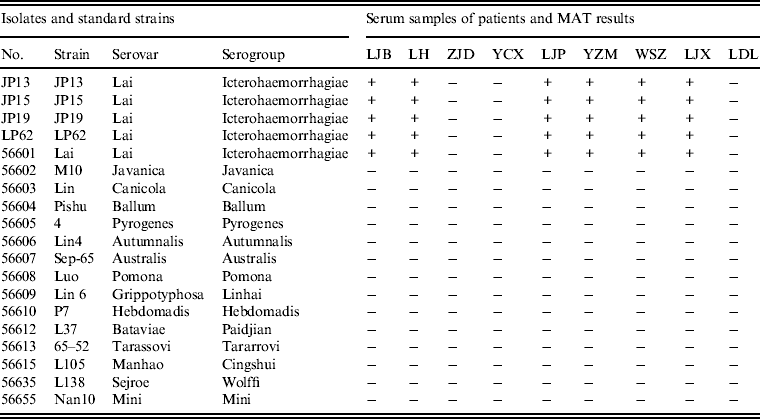
MATs were performed using serum samples from patients with Chinese reference strains belonging to 15 serovars in 15 serogroups of pathogenic Leptospira spp., including the four leptospiral isolates. Of the nine serum samples from patients tested by MAT, six had agglutinating antibodies against isolates JP13, JP15, JP19 and LP62 and reference strain 56601, but not reference strains belong to other serogroups (Table 3).
Table 3. Results of microscopic agglutination tests (MATs) for anti-Leptospira antibody detection in patients from the epidemic region

+, Positive; –, negative.
Confirmation of leptospiral strains
In order to confirm that the four strains of isolates were pathogenic leptospires prior to MLVA analysis, we performed G1/G2 primer-based PCR The results showed that an anticipated 285 bp fragment was successfully amplified in all four isolates and the positive control of L. interrogans serovar Lai strain 56601, but no fragment was detected from the negative control, which confirmed that the four strains of spirochaetes were pathogenic leptospires.
MLVA patterns of leptospiral isolates
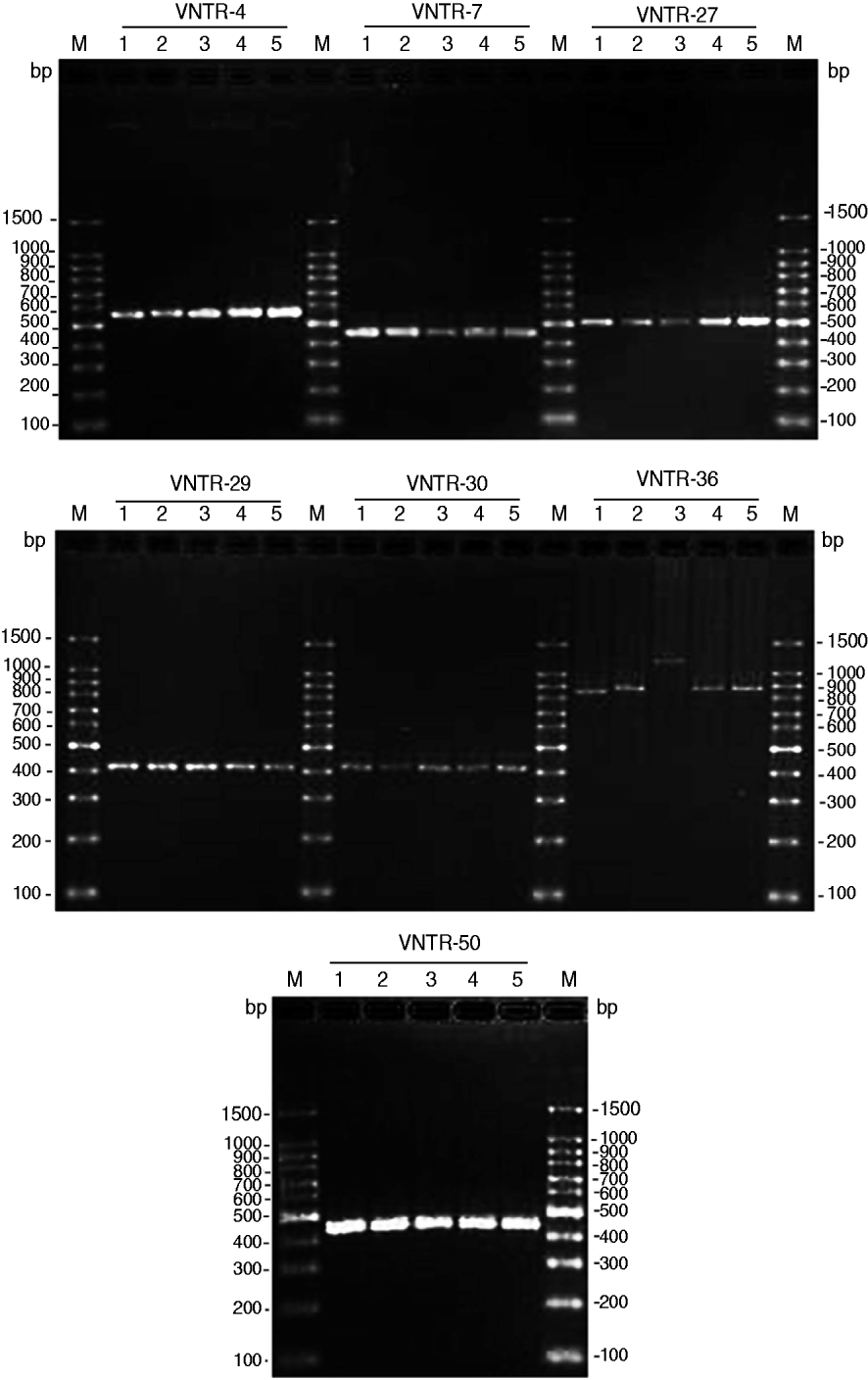
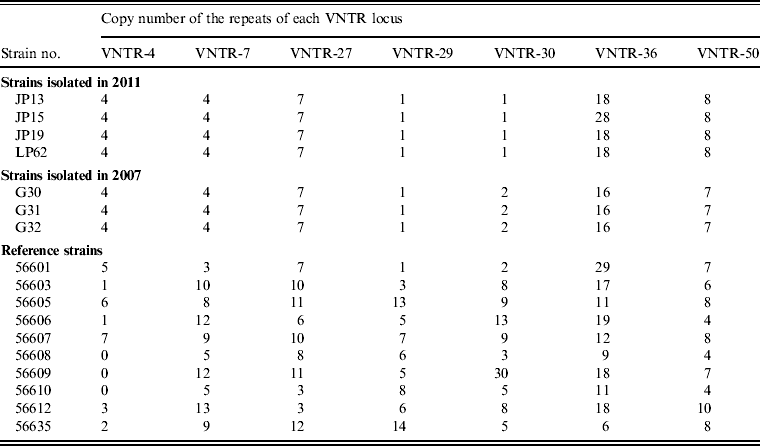
Seven VNTR loci-based primers were used to amplify the chromosomal DNA of leptospiral isolates, and all seven loci were successfully amplified from the four isolates (Fig. 1). The MLVA pattern showed that the four isolates produced the same size of PCR fragment at the same locus, except for strain JP15 at the VNTR-36 locus. The copy number of the repeats of each VNTR locus was deduced from the band sizes of the amplified products. This size was then converted into an allele designation. Among all the isolates, including the four in the present study, the copy numbers of the four isolates were most related to three strains isolated from Rattus tanezumi in Rongjiang county in 2007 as well as the reference strains; moreover, differences in copy numbers for the four isolates and the three strains isolated in 2007 at loci VNTR-30, VNTR-36 and VNTR-50 were also found (Table 4). Further, when compared to the ten reference strains, the copy numbers of the four isolates as well as the three strains isolated in 2007 were most related to reference strain 56601, belonging to serogroup Icterohaemorrhagiae, and differences in copy numbers for the four isolates and strain 56601 at loci VNTR-4, VNTR-7, VNTR-30, VNTR-36 and VNTR-50 were also found.

Fig. 1. PCR products from the seven selected variable number tandem repeat (VNTR) loci of four leptospiral strains isolated from Jinping and Liping counties, Guizhou province. PCR products underwent electrophoresis through a 2% agarose gel. M, 100-bp DNA ladder plus; 1, serovar Lai strain 56601; 2, Leptospira isolate JP13; 3, Leptospira isolate JP15; 4, Leptospira isolate JP19; 5, Leptospira isolate LP62.
Table 4. Results obtained using MLVA for leptospiral isolates and reference strains

MLVA, Multiple locus variable-number tandem repeat analysis, VNTR, variable-number tandem repeat.
Cluster analysis
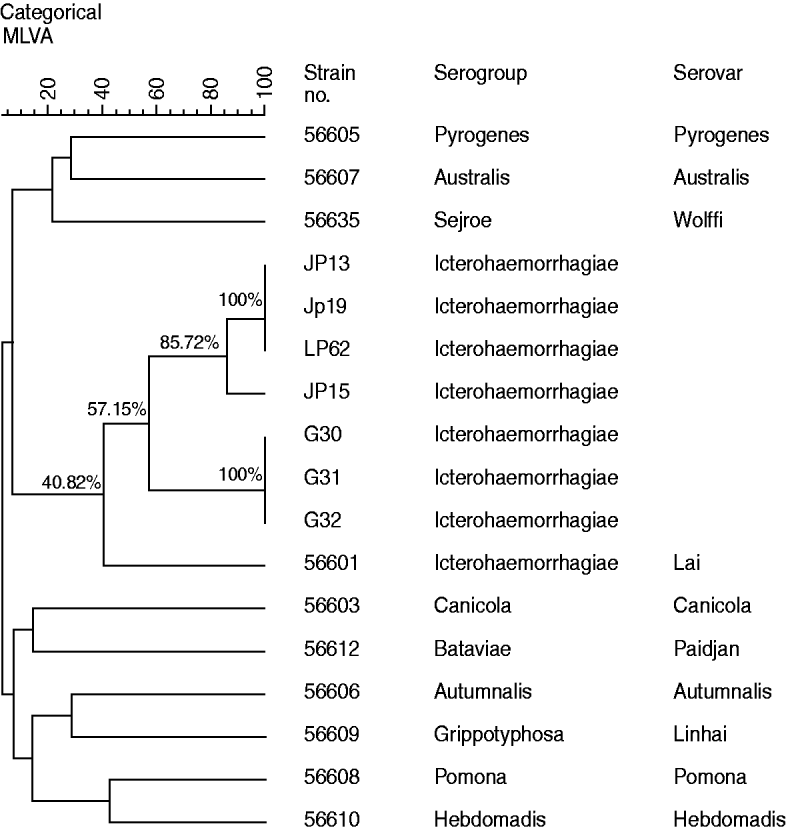
Cluster analysis (Fig. 2), using MLVA data of the four strains isolated from A. agrarius in 2011, three strains isolated in 2007and ten reference strains representative of the ten common epidemic serogroups, suggested that all four leptospiral isolates including the three strains isolated in 2007 and reference strain 56601 formed one clade, which indicated that all the isolates are most closely related to Leptospira serovar group Icterohaemorrhagiae serovar Lai strain 56601. At the same time, the diversity of leptospires isolated from Guizhou province was also revealed by cluster analysis. The isolates JP15 and the remaining strains (including JP13, JP19, LP62) isolated in 2011 clustered at 85·72%, while the three strains isolated from R. tanezumi in 2007 clustered at 100%. Moreover, the four strains isolated from A. agrarius in 2011 and the three strains isolated from R. tanezumi in 2007 only clustered at 57·15%, whereas the seven isolates from Guizhou province and reference strain 56601 only clustered at 40·82%.

Fig. 2. Dendrogram of multiple locus variable-number tandem repeat analysis (MLVA) profile showing Leptospira strains isolated in Guizhou province and ten reference strains..
DISCUSSION
Guizhou province is an old focus of leptospirosis in China [Reference Guo, Deng and Li28–Reference Li30]. Liping and Jinping counties, in Qiandongnan prefecture, were high-incidence regions. For example, 14126 human leptospirosis cases with 534 deaths were reported in Qiannan prefecture from 1958 to 2005. Investigation on the epidemiology of leptospirosis in Liping county, in southeast Guizhou, reveals that a total of 127 leptospirosis cases with 28 deaths were reported from 2001 to 2008 [Reference Yang13]. According to the China National System for Disease Control and Prevention, 12 human leptospirosis cases with one death were reported in Guizhou in 2011. However, these reported cases were only diagnosed clinically, and the source of infection and the characteristic of epidemic bacteria remain unclear.
Rodents are important mammalian reservoirs of Leptospira spp. [Reference Guerra6, Reference Meerburg, Singleton and Kijlstra7, Reference Meerburg, Singleton and Kijlstra31]. Previous studies revealed that A. agrarius is a very important reservoir of leptospirosis with a carriage rate of 7·36%, and accounts for 95·84% of all checked rodents. The geographical distribution of the host animals in the local area is closely associated with the aggregate distribution of cases [Reference Yang and Mo29]. However, few studies on carriage status have been reported in recent years. According to data from China National System for Disease Control and Prevention, the peak incidence is from September to October in the epidemic area; therefore, we performed field mice surveillance and leptospiral carriage status assessment from September to October 2011. The results showed that three strains of leptospires (JP13, JP15, JP19) were isolated from A. agrarius in Jinping county, and one strain (LP62) in Liping county. These strains were typed as Leptospira serogroup Icterohaemorrhagiae which is consistent with the serological detection results by MAT for patients from the epidemic region. MLVA has been proposed for the typing of L. interrogans strains [Reference Salaün32]. The discriminative power of the method in L. interrogans depends on the MLVA loci. It is useful for identifying serovars or for discriminating strains within the same serovar [Reference Salaün32]. Zhang et al. showed that the seven loci-based MLVA used in this study was a reliable method for the typing of leptospiral strains in China [Reference Zhang21].
In the present study, the four strains of L. interrogans isolated from A. agrarius in the high-incidence region were typed as Leptospira serogroup Icterohaemorrhagiae. Our results indicate that A. agrarius is the main carrier of Leptospira in Jinping and Liping counties, and serogroup Icterohaemorrhagiae may be the epidemic serogroup. Moreover, by performing MLVA pattern comparison and cluster analysis of leptospires isolated in different regions and different hosts in Guizhou province, the genetic diversity and relationships among the isolates were clearly described. Our results will contribute to the control and prevention of leptospirosis in these localities.
ACKNOWLEDGEMENTS
This work was supported by Guizhou Province Government Special Funds for Outstanding Scientific and Technological Talent ((2010) 90). We acknowledge the contribution of the Jingping and Liping County CDC for field mice trapping.
DECLARATION OF INTEREST
None.