INTRODUCTION
Bartonella spp. are Gram-negative facultative intracellular bacteria that are highly adapted to their mammalian reservoir hosts, in which they cause chronic relapsing intraerythrocytic bacteraemia [Reference Pulliainen and Dehio1]. Bartonella spp. have been associated with an increasing variety of human diseases, such as trench fever, Carrión's disease, bacillary angiomatosis, endocarditis, cat scratch disease, meningitis and neuroretinitis [Reference Chomel, Boulouis and Breitschwerdt2]. Several Bartonella spp. have been identified as zoonotic or potentially zoonotic. A variety of mammalian species, such as rodents, ruminants and companion animals, serve as reservoirs for several zoonotic species of Bartonella. In dogs (Canis familiaris), at least eight Bartonella species are known to be pathogenic [Reference Perez3]. Although primarily associated with non-clinical bacteremia in domestic cattle (Bos taurus) and wild ruminants, Bartonella bovis infection has been associated with endocarditis in cows in France and in the USA suggesting a role as an animal pathogen [Reference Erol4, Reference Maillard5]. Given its medical importance, much research has focused on known and suspected arthropod vectors. Blood-sucking arthropods, such as fleas (Siphonaptera), lice (Phthiraptera) and ticks (Acari), are potent vectors for the transmission of Bartonella spp. [Reference Reis6].
The deer ked (Lipoptena cervi ) is a common haematophagous ectoparasite of cervids, especially moose (Alces alces) [Reference Kaitala7]. Since the deer ked invaded Finland 50 years ago, its distribution has rapidly spread northwards to the southern parts of Finnish Lapland [Reference Kaitala7, Reference Valimaki8]. This rapid increase in the deer ked population appears to be correlated with high moose densities [Reference Kaitala7, Reference Paakkonen9]. When an adult deer ked attaches to a host, it drops its wings and remains usually for the rest of its life on the same host [Reference Haarløv10]. Following a blood meal, the female gives birth to larvae, which immediately pupate during the autumn and winter. In Finland, pupae drop to the ground or snow and hatch in the following late summer or early autumn. Emerged winged, unfed adults have a short flying period to seek a suitable host during the late summer and autumn [Reference Kaitala7, Reference Haarløv10, Reference Hackman11] (Fig. 1). A prerequisite for the deer ked to act as a successful Bartonella vector is vertical transmission from the mother to its progeny and transstadial transmission from the pupa to the adult. Deer keds in different developmental stages, except the winged adult stage, collected in Germany [Reference Dehio, Sauder and Hiestand12], France [Reference Halos13–Reference Rolain15] the USA [Reference Reeves16] and Norway [Reference Duodu17] have been shown via PCR or culture to harbour Bartonella spp. However, the question whether the deer ked is a competent vector for the transmission of Bartonella spp. remains open. Although Bartonella spp. have been isolated or amplified from both pupae and adult wingless deer keds, transstadial transmission remains to be demonstrated.

Fig. 1. Schematic overview of the deer ked life cycle.
Given that deer keds incidentally bite humans [Reference Härkönen18] there is a potential risk for occasional transmission of B. schoenbuchensis. In fact, B. schoenbuchensis has been considered a possible aetiological agent of deer ked dermatitis [Reference Dehio, Sauder and Hiestand12], which resembles cat scratch disease, usually a self-limiting febrile illness caused by B. henselae. In Finland, since the 1970s, there have been an increasing number of people suffering from recurrent and occasionally long-lasting dermatitis associated with deer ked bites [Reference Härkönen18]. Although L. cervi does not reproduce on humans, the infestation is a nuisance for people who participate in outdoor activities during late summer and early autumn.
Based on the heavy deer ked parasitism on Finnish moose [Reference Paakkonen9], we hypothesized that the same Bartonella spp. could be demonstrated in deer keds and moose in Finland. The aim of this study was threefold: first, to investigate whether Finnish deer keds carry bartonellae, and if they do, to determine the molecular diversity, prevalence and geographical distribution of the identified Bartonella spp.; second, to seek evidence of possible vertical and/or transstadial transmission of Bartonella spp. in deer keds; and third, to investigate whether Finnish moose and deer keds carry the same Bartonella spp., which would further support the role of the deer ked in the transmission of Bartonella spp.
METHODS
Sample collection
Deer ked pupae were collected from the surface of the snow on moose bedding sites in late winter 2007. A total of 156 pooled pupae samples representing 1154 pupae were screened: 54 pooled samples (2–40 pupae in each pool) and 102 individual pupae. The unfed winged adults (59 pools of 2 keds/pool; n = 118) were collected in autumn 2008 by walking in the forest and capturing deer keds that attached to the investigators. The pupae and the adults were stored at −70°C and −20°C, respectively, until processing. Moose blood samples (n = 8) were collected by Finnish hunters during the hunting season in 2012. After shooting the moose, 7–10 ml of whole blood from the jugular vein or thoracic cavity were collected from each moose into 10 ml EDTA tubes.
Homogenization and DNA extraction
The deer ked pupae and adults were homogenized in porcelain mortars with sterile sand in Dulbecco's phosphate buffered saline (DPBS) supplemented with 0·2% (w/v) of bovine serum albumin. For the pooled pupae, DPBS was supplemented with antibiotics (10 U/ml penicillin and 0·1 mg/ml streptomycin). The individual pupae and the adults were surface-sterilized before homogenization with 75% EtOH for 5 min with no antibiotics. The DNA of the pooled pupae was extracted with Tripure isolation reagent (Roche Diagnostics GmbH, Germany) according to the manufacturer's instructions, except that DNA was eluted in Tris EDTA buffer (pH 8·0). DNA extraction from individual pupae and adults as well as moose blood was performed using DNeasy Blood & Tissue kits (Qiagen, Germany).
PCR analyses
Detection of Bartonella spp. was performed using two different PCR strategies. All samples were initially screened with a nested PCR method targeting the 16S rRNA gene of Bartonella spp. as described previously [Reference Garcia-Esteban19]. To improve the sensitivity of the PCR method, nested primers were designed using Primer3 software (Whitehead Institute for Biomedical Research, MIT, USA) based on obtained sequences and those from the GenBank nucleotide sequence database. Amplification was performed in a 25 μl final volume reaction containing 2 mm MgCl2, 1x Taq buffer with KCl, 0·2 mm dNTP, 0·4 μ m of each primer (16Si-F: 5′-CAG CTC GTG TCG TGA GAT GT-3′ and 16Si-R: 5′-CAG AGT GCA ATC CGA ACT GA-3′), 2·5 U recombinant Taq polymerase (Fermentas, USA) and 2·5 μl DNA template. The primers amplified a 250 bp amplicon. The first round of the nested PCR was performed as described previously [Reference Garcia-Esteban19], with the exception that the number of the cycles was decreased from 45 to 40 and 2·5 U of TrueStart Taq polymerase (Fermentas) was used. The second round was performed with 1 μl of the first round product in a total volume of 50 μl, and the annealing temperature was set to 54°C with a total of 30 cycles. To avoid nested PCR contamination, sample preparation, DNA extraction, PCR preparation and nested PCR amplification and analysis were performed in separate rooms. For species identification of the positive samples, PCR was used to produce amplicons from the beta subunit of the RNA polymerase gene (rpoB). The following rpoB oligonucleotides prAPT0244 (5′-GATGTGCATCCTACGCATTATGG-3′) and prAPT0245 (5′-AATGGTGCCTCAGCATATAAG-3′) were used in a previously described protocol [Reference Oksi20]. For the second round of nested PCR, 1 μl of PCR product of the first round was used as template. DNA from a Finnish human patient with B. quintana-associated endocarditis was used as a positive PCR control in both methods [Reference Vento21]. Distilled water was used as negative control. PCR products were analysed by 2% agarose gel electrophoresis under UV exposure. The sizes of the amplicons were determined by comparison with the molecular weight of a standard marker (SM0323; Thermo Fisher Scientific, Germany).
Sequence and phylogenetic analyses
All PCR products were directly sequenced and unique sequences deposited into GenBank. The accession numbers for the 16S rRNA sequences of the pupae are JN542708–JN542712 and JN542713 for the winged adults. The accession numbers for the rpoB sequences are KJ739719–KJ739723 (two from pupae and three from moose). Bartonella spp. annotation was performed by comparing similarities with other sequences deposited in the GenBank database using the Basic Local Alignment Search Tool (BLAST v. 2.0, McAfee, USA). Obtained and selected sequences listed in the GenBank nucleotide sequence database were aligned with BioEdit (Ibis Biosciences, USA) [Reference Hall22] and the phylogenetic tree was constructed based on the rpoB gene using the DNAML program from the PHYLIP package [Reference Felsenstein23].
RESULTS
We collected a total of 1154 pupae from 13 different locations within the deer ked distribution area and 118 adult winged keds from a single location (Pulkkila). All pupae samples were analysed either in pools (54 pools of 2–40) or individually (102 pupae). Fifty-nine pools (2 keds per pool) of adult, winged deer keds were screened. Using 16S rRNA Bartonella genus-specific primers, Bartonella DNA was successfully amplified from 12/156 (7·7%) deer ked pupae and 1/59 (1·7%) deer ked pools from eight different locations (Fig. 2). All locations where more than three pools of pupae were collected contained at least one sample that was positive for Bartonella (Table 1, Fig. 2). All samples from locations with three or less pools remained negative (Pörtom 0/1, Juva 0/2, Nilsiä 0/1, Kontiolahti 0/2, Laukaa 0/1, Leppävirta–Heinävesi 0/3 samples). Pulkkila, the location with the positive winged adult, is close to the northernmost limit of the deer ked distribution area [Reference Kaunisto24] (Fig. 2).

Fig. 2. Map of Finland depicting the geographical locations of Bartonella-positive samples obtained from deer keds (1–8) and moose (*, #). 1, Siikainen; 2, Yläne; 3, Mynämäki; 4, Kuhmoinen; 5, Kitee; 6, Kuopio; 7, Lemi; 8, Pulkkila; * Liperi; # Hyvinkää.
Table 1. Geographical origin of the Lipoptena cervi samples and the prevalence of Bartonella infection based on PCR and DNA sequencing
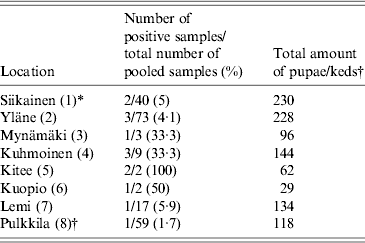
* Numbers in parentheses in the first column indicate numbering of locations as in Figure 2.
† All winged adult keds were collected at one location, Pulkkila.
In addition, eight EDTA blood samples were collected from free-ranging moose for Bartonella testing. Five samples were collected in Liperi and three in Hyvinkää (Fig. 2). All eight moose samples were Bartonella PCR positive. The overall prevalence of Bartonella DNA-positive pupae in the pools was ~0·7% (7/1052), assuming there was at least one Bartonella DNA-positive pupa per one positive pool. In addition, 5/102 (4·9%) individually processed pupae collected from three locations were positive and 1/59 (1·7%) of the adult winged deer keds harboured Bartonella DNA; likewise, if assumed that only one ked in a positive pool of two carried Bartonella, 1/118 (0·8%) of the adults were positive (Table 1).
The amplified product ranged from 200–250 bp in length. Sequence analysis confirmed the presence of Bartonella DNA in the screened deer ked and moose samples but identification at the species level was not possible due to high level of sequence conservation. However, two distinct PCR sequence groups, identical either to B. bovis or B. schoenbuchensis, were identified, based on one signature nucleotide substitution. Five deer keds (from which DNA samples were available) and all moose samples that were Bartonella-positive using the 16S rRNA PCR were subsequently tested for the rpoB gene, of which 4/5 and 8/8 were positive. A maximum-likelihood phylogenetic tree was generated from the alignment of the rpoB fragments from all deer ked and moose sequences described as well as representatives of Bartonella isolates and Bartonella spp. deposited in Genbank (Fig. 3). The phylogeny demonstrated that there are two Bartonella lineages in deer keds in Finland: the majority of the sequences from deer keds (10/13, 76·9%) and moose (5/8, 62·5%) clustered with B. schoenbuchensis, whereas 23% (3/13) of the sequences from deer keds and 37·5% (3/8) from moose clustered within another clade more closely related to B. bovis. Sequence analysis demonstrated no geographical clustering of either of the clades.
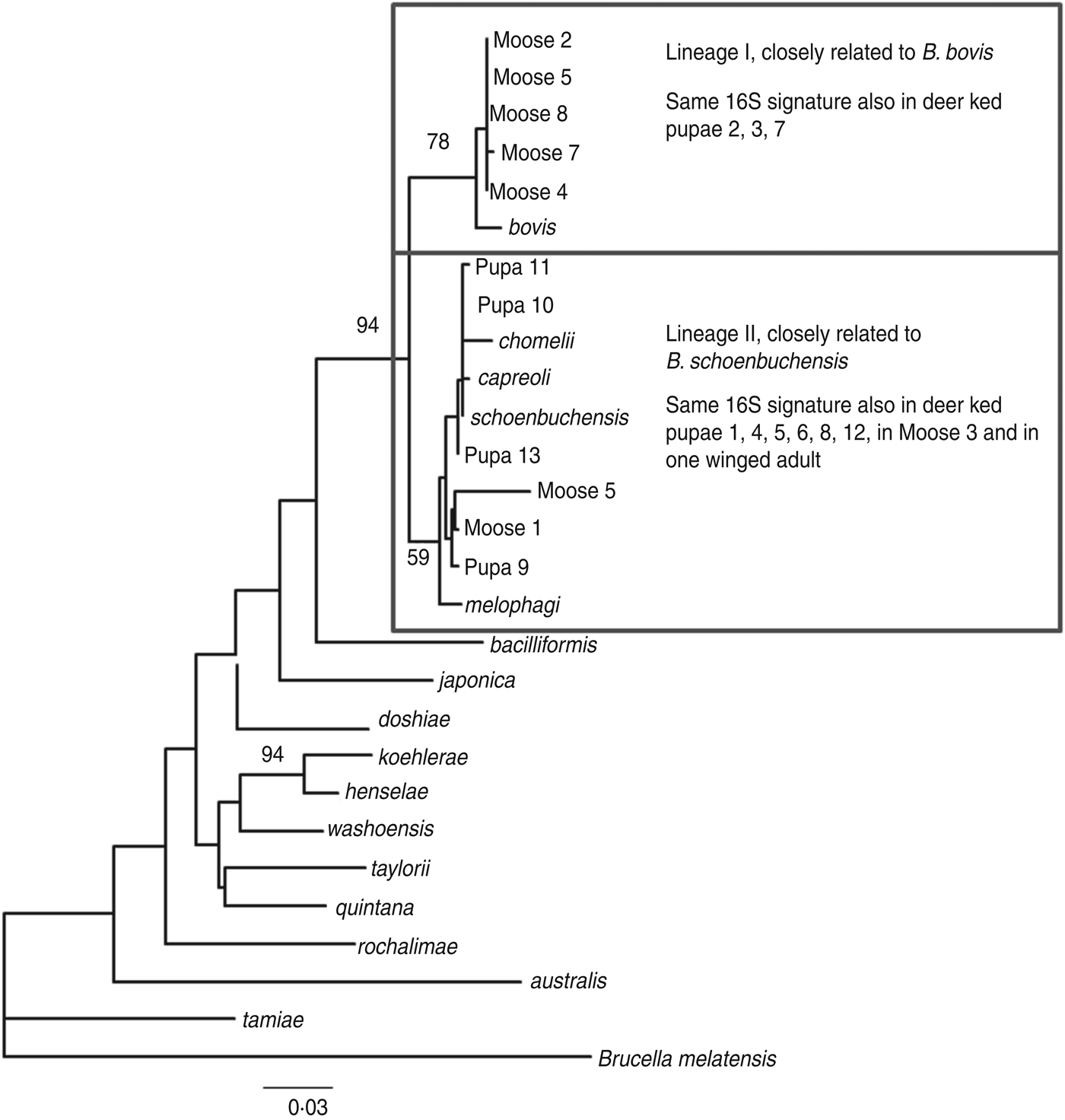
Fig. 3. Maximum-likelihood phylogenetic tree based on partial nucleotide sequences of the rpoB gene, estimated using the DNAML program from PHYLIP. Bootstrap support values are given for the major nodes including sequences derived in this study. Clustering pattern of additional samples, from which 16S sequences were derived, is indicated in the boxes. The 16S signature is A at position 16 of the 182 nt fragment for lineage I, and C for lineage II. The scale bar indicates evolutionary distance of 0·03 nt per position in the sequence.
DISCUSSION
In the present study, we provide the first molecular evidence for Bartonella spp. DNA in an unfed winged adult deer ked, indicating that this organism or at least its DNA could survive in the deer ked from one life stage to another (transstadial transmission).
Bartonella spp. are Gram-negative bacteria that infect erythrocytes and endothelial cells [Reference Pulliainen and Dehio1]. Because of the ability of this organism to reside within red blood cells of diverse hosts, there is a considerable opportunity for the uptake of these bacteria by several haematophagous arthropods. Bartonella spp. are transmitted by lice, fleas, sandflies (Phlebotominae) and ticks [Reference Reis6]. However, Bartonella spp. have also been detected by PCR or culture from several other arthropods, such as deer keds [Reference Dehio, Sauder and Hiestand12, Reference Duodu17]. Previously, B. schoenbuchensis-like bacteria were detected in deer keds collected from cervids. However, all these keds had dropped their wings and therefore were likely to have started consuming blood [Reference Dehio, Sauder and Hiestand12, Reference Halos13, Reference Reeves16, Reference Matsumoto25]. On the other hand, B. bovis was previously reported in the USA [Reference Chang26, Reference Cherry27] and in France [Reference Maillard5]. Recently, researchers in Norway investigated the presence of Bartonella spp. in pools of winged unfed deer ked imagines [Reference Duodu17]. However, Bartonella DNA was not successfully amplified nor could the bacteria be cultured. Thus, our data show for the first time preliminary evidence for transstadial transmission of Bartonella spp. in the deer ked. These results need to be confirmed with larger studies and isolation of viable bacteria.
Based on DNA sequencing, the same two species were identified in both deer keds and moose with practically exact sequences. In a recent study in Norway, two different Bartonella clades were found in deer keds and moose after PCR and sequencing [Reference Duodu17]. One lineage was similar to B. schoenbuchensis, B. chomelii and B. capreoli, and a distinct lineage of Bartonella was observed both inside and outside the deer ked range. We identified one Bartonella lineage similar to the corresponding sequences from B. schoenbuchensis, and a different lineage, closely related to B. bovis, both within the deer ked distribution range.
Although the number of blood samples was low, our data suggest a high prevalence of Bartonella infection in moose in Finland. Similarly, high prevalence of Bartonella spp. was previously reported in moose blood (70%) collected in Norway [Reference Duodu17]. In that study, the prevalence of Bartonella infection in moose in the deer ked zone was higher than in moose from deer ked-free areas (70% vs. 37%), which suggested that the deer ked infestation may have been responsible for the high prevalence of Bartonella infection. In our study, no geographical cluster was observed in the sequence analysis. However, we were unfortunately not able to test blood samples from moose outside the deer ked zone.
Recently, the prevalence and distribution of Bartonella infection were investigated in cattle in five different countries (Kenya, Thailand, Japan, Georgia, Guatemala) and in buffaloes (Bubalus bubalis) from Thailand [Reference Bai28]. The authors reported that the prevalence of Bartonella infection varied extremely across the regions studied, from 0% to 90%. Future epidemiological studies with a larger number of moose blood samples are necessary to determine if geographical differences in Bartonella prevalence in moose also occur in Finland.
Since its first association with endocarditis in 1993, Bartonella infection has become an important cause of culture-negative endocarditis in humans [Reference Chomel, Boulouis and Breitschwerdt2]. Endocarditis has also been reported in cats (Felis catus) due to B. henselae as well as in dogs due to B. vinsonii subsp. berkhoffii and B. koehlerae. To date, it is not known whether chronic bacteremia with B. bovis and B. schoenbuchensis has any impact on the health of the moose. In Finland, up to 17 000 keds can be found on adult moose bulls while counts are lower on cows and calves [Reference Paakkonen9]. Based on a Finnish study, moose in deer ked-free areas did not show better indices of health compared to infested animals [Reference Paakkonen29]. Although ruminant-infecting Bartonella are assumed to be of minimal pathogenicity, B. bovis has been detected in diseased heart valves of cows by PCR [Reference Maillard5] and also in a cow diagnosed with B. bovis endocarditis by PCR and DNA sequencing, serology and culture [Reference Erol4]. Given the close association between the deer ked and its ruminant host, as well as the incidental infestation of humans with this arthropod, the deer ked may serve as a competent vector for the transmission of Bartonella within ruminants and to people.
In conclusion, our data support the potential of the deer ked for vector competence of Bartonella spp. The present study also demonstrates that adult winged deer keds may harbour bartonellae. Two different lineages, B. bovis and B. schoenbuchensis, were detected widely throughout the distribution range of deer keds and in moose. It remains to be determined if the geographically widespread distribution of deer keds is responsible for the Bartonella bacteraemia detected in moose in Finland.
ACKNOWLEDGEMENTS
We thank the numerous hunters and other volunteers for their assistance in collecting the moose samples, pupae and adult deer ked specimens for screening. We thank Tytti Manni for technical assistance and the Hospital District of Helsinki and Uusimaa (HUS-EVO TYH2008309 and TYH2011305) for financial support.
DECLARATION OF INTEREST
None.







