INTRODUCTION
Microsporidia are single-celled, spore-forming obligate intracellular parasites which, prior to 1985, were only sporadically reported in humans. They have since emerged as a major cause of opportunistic infections associated with persistent diarrhoea and weight loss in persons with AIDS [Reference Didier1] and organ transplant patients [Reference Lanternier2–Reference Sing4]. While still uncommonly reported in immunocompetent persons, intestinal microsporidiosis is increasingly being identified in children, the elderly, and travellers [Reference Fournier5–Reference Wichro8]. In healthy persons, disease is characterized by self-limiting diarrhoea usually of ⩽1 month's duration [Reference Didier1]. The incubation period for microsporidiosis remains unknown.
Although there are more than 1200 species of microsporidia, most infections in humans are caused by two species: Enterocytozoon (E.) bieneusi and Encephalitozoon intestinalis [Reference Mathis, Weber and Deplazes9]. E. bieneusi has been identified in both immunocompromised and healthy persons and has also been isolated from a wide range of animal hosts including cattle, pigs and a variety of birds [Reference Didier6, Reference Bart10, Reference Lobo11]. More than 90 genotypes of E. bieneusi are recognized, some of which are associated with a single host and others with ⩾2 hosts [Reference Santin and Fayer12]. Possible modes of transmission include person-to-person, zoonotic transmission, inhalation of contaminated aerosols and ingestion of contaminated food and water [Reference Didier6]. Only one outbreak of microsporidiosis, associated with waterborne transmission, has been reported in the literature [Reference Cotte13].
Previously, microsporidia research in Sweden had focused on HIV-positive patients [Reference Svedhem14, Reference Svedhem15]. In contrast, only one case of E. bieneusi infection in an immunocompetent person has been reported [Reference Svenungsson16]. The case-patient was an HIV-negative man who had not travelled abroad in the previous 12 months and was identified as part of a 1-year prospective study to identify enteropathogens in adults with diarrhoea and healthy control subjects [Reference Svenungsson17]. Of 417 patients with diarrhoea found to be shedding enteropathogens, only this one microsporidiosis case was identified; out of 203 controls none were found to have microsporidia.
We describe a large foodborne outbreak of microsporidiosis associated with a hotel in central Sweden. On 5 November 2009, the regional public health authority in Värmland County was informed of more than 100 cases of gastrointestinal illness in 471 attendees of a 1-day conference held at a hotel on 23 October 2009 (group A). A second group of visitors (n=15, group B) attended a separate meeting at the hotel on the same day but reported no illness. Staff members working on 23 October (n=39) had also eaten the same foods on that day and two reported illness after the event. In collaboration with the regional public health and environmental health authorities, the Swedish Institute for Communicable Disease Control (SMI) initiated an investigation in order to determine the mode and vehicle of transmission, establish the magnitude of the outbreak, and identify the aetiological agent.
METHODS
Laboratory investigations
Outbreak samples
All persons who reported illness were asked to submit a stool sample. The county microbiology laboratory investigated 95 samples for bacterial enteropathogens (Salmonella, Shigella, Campylobacter, enterohaemorrhagic Escherichia coli, Vibrio, Plesiomonas, Aeromonas) and intestinal parasites (Entamoeba spp., Giardia, Isospora, Cyclospora, Cryptosporidium).
Of these initial stool samples, only eight (formalin-fixed) samples remained for further testing and were sent to SMI's parasitology reference laboratory where they were investigated by light microscopy for Entamoeba spp., Dientamoeba, Giardia, Cryptosporidium, Blastocystis and Cyclospora. Subsequent examination for the presence of microsporidian spores was performed by light microscopy after modified trichrome staining [Reference Weber18] and monoclonal antibody immunofluorescence assay (Bordier Affinity Products SA, Switzerland). DNA was extracted directly from stool specimens by using the QIAamp DNA mini kit (Qiagen, Germany) after an initial disruption of the spores with a Mini-BeadBeater (Biospec Products Inc., USA). Amplification of the ITS region was performed by using the primers for nested PCR described by Buckholt et al. [Reference Buckholt, Lee and Tzipori19]. Amplicons from all PCR-positive samples were directly sequenced in both directions. The BLAST tool was used to compare the nucleotide sequences with sequences in the GenBank database.
A further three samples from persons still experiencing symptoms in late November were analysed for the presence of norovirus, rotavirus and adenovirus. These samples were subsequently sent to SMI's virology laboratory where they were tested for astrovirus, rotavirus, adenovirus, norovirus and sapovirus. They were also later investigated for microsporidia at SMI's parasitology laboratory.
DNA from all samples available at SMI (n=11) were sent to the reference parasitology laboratory at Statens Serum Institut (SSI) in Copenhagen for investigation of E. bieneusi and Encephalitozoon spp. by real-time PCR [Reference Verweij20].
Prevalence study samples
In June 2010, all members of the same professional group as group A who had not attended the conference on 23 October (n≈70) were contacted and invited to participate in a prevalence study. They were asked to provide a stool sample and complete a short paper questionnaire regarding sex, age, travel history during the previous 6 months, and gastrointestinal illness during the previous 4 weeks. DNA from samples was extracted and amplified as described above. Ethical approval was obtained from the ethical review board at Karolinska Institute, Solna, Sweden.
Epidemiological investigation
We conducted a retrospective cohort study which included the three identifiable groups who ate at the hotel on 23 October: group A, group B and the hotel staff who worked on that day. All participants were asked to complete a web-based questionnaire (Artisan Global Software, Sweden; www.artologik.com). For group A and the hotel staff, the link to the questionnaire was sent by email on 18 November. For group B, only postal addresses were available so all 15 persons were sent a letter with the link to the questionnaire together with a paper copy. The questionnaire asked about date and time of illness onset, type and duration of symptoms, foods consumed during the entire day (coffee breaks, lunch, dinner) and other activities at the hotel (overnight stay, use of spa facilities).
A case-patient was defined as a person who ate at the hotel on 23 October 2009, and subsequently developed at least one of the following symptoms: diarrhoea, vomiting, abdominal pain or nausea. An onset date was also required.
We analysed the data using Stata v. 10 (StataCorp, USA). Food-specific attack rates (AR), relative risks (RR) and 95% confidence intervals (CIs) were calculated for all exposures. A Fisher's exact P value of <0·05 was considered significant. Respondents who answered ‘do not know’ were excluded from the analysis of that particular exposure. For those persons who stated that they ate each meal and answered yes to one or more of the food items for that meal and who left all other answers blank, the missing values were coded as ‘no’. For all others, missing values were retained and these persons were excluded from the analysis. The univariate analyses of the individual food items served during the morning coffee break and lunchtime meals were restricted to persons who attended those meals. To control for confounding, we performed logistic regression. For both the morning coffee break and lunchtime meals, food items that were significantly associated with illness (P was <0·05) and were considered to be biologically plausible sources of infection were fitted into a model together with sex and age group.
Environmental investigation
The local Environmental Health Officer (EHO) inspected the hotel kitchen on several occasions and interviewed the staff. In addition to evaluating the temperature controls and observing general hygiene practices, the EHO investigated the food handling and storage procedures for the foods served on 23 October. No leftover food samples were available for microbiological testing.
RESULTS
Laboratory investigations
Outbreak samples
Initially, 95 stool samples were obtained. All samples were negative for enteropathogenic bacteria and intestinal parasites at the county laboratory. The three additional samples from patients with continuing symptoms were negative for norovirus, rotavirus and adenovirus (Table 1).
Table 1. Summary of results from laboratory investigations of stool samples submitted following the hotel visit on 23 October 2009

SMI, Swedish Institute for Communicable Disease Control, Solna, Sweden; SSI, Statens Serum Institut, Copenhagen, Denmark.
* E. bieneusi was detected by using light microscopy (chromotrope staining) in combination with an immunofluorescence assay.
† Of those samples positive for E. bieneusi at SMI, all six were genotype C: GenBank accession no. AF101199.
‡ SSI detected one further case by real-time PCR among the eight initial, formalin-fixed samples. This sample was not genotyped.
None of the eight stool samples sent to SMI were positive for Giardia, Cryptosporidium, Cyclospora, Dientamoeba, Entamoeba spp., or Blastocystis. Similarly, the faecal samples from the three persons with persisting symptoms in late November were all negative for enteropathogenic viruses at SMI.
On 17 December, 3/8 initial samples received at SMI were found to contain microsporidia spores by microscopy. An immunofluorescence assay using monoclonal antibody identified the species in all three samples as E. bieneusi. This was further confirmed by PCR. Two of the three samples from late November were positive for E. bieneusi by microscopy, immunofluorescent staining and PCR; the remaining sample was positive only by PCR. Thus, a total of six samples were positive for E. bieneusi. Sequencing of these PCR-positive samples revealed that all six had sequences corresponding to the previously described genotype C, GenBank accession no. AF101199 (Table 1).
Real-time PCR performed by SSI detected E. bieneusi in 4/8 initial stool samples and all three of the samples from late November. This gave a total of 7/11 positive samples. No samples were positive for Encephalitozoon spp.
Prevalence study samples
A total of 19 control group persons provided stool samples and completed the questionnaire. All samples were negative for microsporidia. The age and gender distribution of these persons was comparable to that of the outbreak cohort (95% female, median age 56 years) and none had experienced gastrointestinal illness in the previous 4 weeks.
Epidemiological investigation
In total, 450/525 (86%) persons completed the questionnaire and 135 met the case definition (AR 30%). Of the staff, only 3/39 (AR 8%) reported illness while in groups A and B the ARs were 33% (130/397) and 14% (2/14), respectively. The median age of the case-patients was 52 years (range 20–65 years) and most (86%) were female; these figures were similar to those of the cohort as a whole (median age 52 years, 84% female).
Onset dates of the 135 case-patients ranged from 23 October to 13 November, with a peak on 2 November (Fig. 1). The distribution of case-patients over time indicates a point-source outbreak but is broad, reflecting a wide range of incubation periods. The median incubation time between the date of the conference and illness onset in case-patients was 9 days for all case-patients (range 0–21 days) and 7 days (range 3–15 days) for the four microbiologically confirmed case-patients (Table 2).
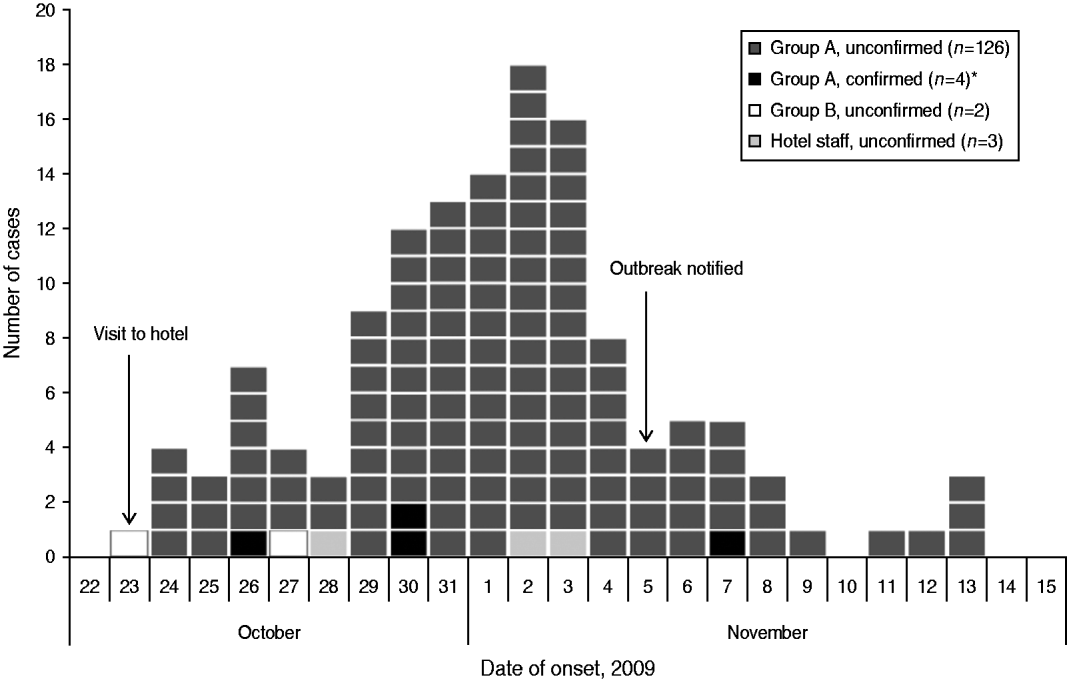
Fig. 1. Distribution of cases (n=135) by date of symptom onset during an outbreak of E. bieneusi, Sweden, October–November 2009. * Three of the microbiologically confirmed case-patients did not return the questionnaire and are therefore not shown in the figure. Two of these three case-patients became ill on 2 November; an onset date for the remaining case-patient was not available.
Table 2. Summary of symptom distribution, symptom duration and incubation periods in case-patients during an outbreak of E. bieneusi, Sweden, October–November 2009

* Three of the seven microbiologically confirmed case-patients did not return the questionnaire and are therefore not included here.
† These symptoms were reported by case-patients in a free-text ‘other symptoms’ question.
‡ Median and range of symptom duration could only be calculated for those case-patients whose symptoms had ceased at the time of questionnaire completion and who did not have missing data (n=89). For confirmed case-patients, three indicated that they were still ill so no median or range of symptom duration could be calculated.
§ One case-patient developed symptoms on the same day as the event; the incubation time was 8–10 h.
The most common symptoms were abdominal pain (87%), diarrhoea (82%) and nausea (82%) (Table 2). Notably, vomiting was only present in 7% of case-patients. When asked about other symptoms (free-text field), 29 case-patients (21%) reported feeling bloated, 14 (10%) reported suffering from flatulence and 13 (10%) reported experiencing fatigue. None of the case-patients had bloody diarrhoea. The median duration of illness was 6 days (range 1–20 days) although it should be noted that the regional public health authority was notified of case-patients who were still experiencing persistent or recurring symptoms as late as February 2010.
Persons who attended the morning coffee break were found to have a 6·4 times higher risk of illness than those who did not (Table 3). Lunch was also associated with illness (RR 5·7, 95% CI 0·8–38·1, P=0·018). Of the individual food items, the cheese sandwich served during the morning coffee break and the salad served during lunch had the highest RRs: 4·1 (95% CI 1·4–12·2, P=0·001) and 2·1 (95% CI 1·1–4·0, P=0·013), respectively (Table 3). Of the 135 cases, 130 (97%) ate the cheese sandwich and 117 (87%) ate the salad. Additionally, the mashed potatoes and bread were significantly associated with illness. Univariate analyses for food items served during the afternoon coffee break, at dinner, and for other snacks provided throughout the day were also performed (data not shown). Although some of these items were significantly associated with illness, the RRs were <2 and none accounted for more than 80% of cases.
Table 3. Risk of illness by exposure to foods served during a coffee break and lunch on 23 October 2009, Sweden

AR, Attack rate; RR, relative risk; CI, confidence interval.
* Fisher's exact P value.
† The salad contained lettuce, tomatoes, cucumber and sweetcorn.
In a multivariable analysis which included three food items (cheese sandwich, salad, mashed potatoes) as well as sex and age group, only the cheese sandwich and the salad continued to show a significant association with illness: the odds ratio (OR) for the cheese sandwich was highest at 6·57 (95% CI 1·53–28·28, P=0·011) while the OR for salad was 2·49 (95% CI 1·12–5·54, P=0·026).
Environmental investigation
General food-handling practices at the hotel were considered to be good; no lapses in temperature control or general hygiene were identified. Further investigation of the cucumber used in the cheese sandwiches revealed that it was supplied by a wholesaler who had washed, sliced and packaged the product in sealed plastic bags prior to delivery. No further washing of the cucumber slices was performed at the hotel. At the same time, the supplier had also delivered whole, unwashed cucumbers which were used in the salad served at lunch time; these cucumbers were washed and chopped at the hotel. It was subsequently established that cucumber slices left over from making the cheese sandwiches had been added to the salad. Further investigations revealed that the pre-sliced cucumbers had been supplied to the wholesaler by an intermediate supplier who had imported them from Spain. The whole cucumbers used in the salad came from a different intermediate supplier and the country of origin could not be traced.
DISCUSSION
We describe the first identified foodborne outbreak to be associated with the microsporidian E. bieneusi. Our investigations suggest that cucumber slices in both cheese sandwiches and a salad were the most probable vehicle of transmission. Since no leftover food samples were available for testing and because little is known about E. bieneusi in the context of foodborne outbreaks, it is difficult to conclusively implicate this organism as the agent responsible for the outbreak. However, the finding that all six samples available for genotyping were genetically indistinguishable (genotype C) together with the fact that, despite extensive testing, no other organisms were identified in the stool samples strongly suggest that E. bieneusi was the causative agent. Furthermore, the finding that all 19 stool samples from persons belonging to the same professional group who had not attended the event were negative for microsporidia provides additional evidence that the detection of E. bieneusi was not a chance finding. Although these samples were taken 7 months after the event, they nevertheless provide an indication of the prevalence of microsporidia in a population with similar demographic characteristics.
The only outbreak of microsporidia reported in the literature was identified retrospectively by analysis of all samples submitted for microsporidial testing during a 3·5-year period in France; the cause of the outbreak was attributed to drinking water because case-patients lived in the same water distribution area; however, because no analytical epidemiological study was done it is difficult to draw meaningful conclusions from this data [Reference Cotte13]. Nevertheless, food- and waterborne routes of transmission of microsporidia are biologically plausible and are supported by several findings including: environmental resistance of spores, probable resistance to disinfectants, survival of spores in water for extended periods, and the detection of spores in ground [Reference Dowd, Gerba and Pepper21], surface [Reference Dowd, Gerba and Pepper21, Reference Fournier22], and crop-irrigation [Reference Thurston-Enriquez23] waters, and in fresh produce such as raspberries, beans and lettuce [Reference Jedrzejewski24].
We cannot state with certainty how and where the sliced cucumbers were contaminated. Contamination during final preparation at the hotel seems unlikely because the cucumbers were not processed any further but were added directly to the sandwiches. Furthermore, a high contamination dose is suspected (due to high attack rate in a healthy population) which is unlikely to have occurred because preparation of the sandwiches was carried out by an asymptomatic food handler. The sealed bags of cucumber slices had been refrigerated before use so it is improbable that contamination took place during storage. Similarly, contamination during initial processing at the wholesale supplier, although possible, seems unlikely based on the description of the procedures used. The most likely hypothesis of contamination is that it occurred before harvest, either by contaminated manure, manure compost, sewage sludge, irrigation water, runoff water from livestock operations or directly from wild and domestic animals. These potential contamination events are all plausible and consistent with the assumption that the level of contamination must have been high. Unfortunately, because we were unable to trace the cucumbers back to the farm where they were grown, we could not investigate these possible contamination routes further. However, additional information is provided by the genotyping results. While there have been several cases of genotype C identified in humans, predominantly in HIV-negative organ transplant recipients in Europe [Reference ten Hove25, Reference Liguory26], there is only one report on animals in the literature [Reference Sak27]. Thus, while a zoonotic link cannot be ruled out, the involvement of this genotype suggests that the source of contamination in this outbreak was of human (faecal) origin.
While thorough washing of fresh produce remains of utmost importance in preventing foodborne illness and should continue to be emphasized, sometimes washing may be insufficient to remove all pathogens. In this instance, it may have been that the level of contamination was so high that washing was unable to remove enough of the microbial load so as to prevent infection. Alternatively, it may be that microsporidian spores are capable of strong adhesion to, or internalization in, certain types of produce, thereby successfully evading the effects of washing and disinfection. A recent paper by researchers in the USA demonstrated that Cryptosporidium oocysts were capable of strongly adhering to spinach plants after contact with contaminated water and were also internalized within the leaves, thus making washing entirely ineffective [Reference Macarisin, Bauchan and Fayer28].
Several limitations apply to our findings. First, the long incubation period resulted in a substantial delay before the outbreak was recognized and this delayed the start of the epidemiological investigation. As a result, the web-based questionnaire was sent out almost 4 weeks after the event and this may have resulted in recall bias. An additional consequence of the delay was that it was not feasible to enquire about the amounts of foods consumed so we were unable to calculate dose response. This is unfortunate because very little is known regarding infection with E. bieneusi in healthy populations and we strongly suspect that the high variability in the incubation period, spectrum, severity and duration of symptoms could be explained by dose response. Another limitation associated with the delayed recognition of the outbreak was that only 8/95 initial samples remained for extended analysis. Furthermore, these samples had been fixed in formalin ⩾6 weeks before DNA extraction and this may have influenced the low PCR-positivity rate (4/8). In contrast, all three stool samples from late November which had been stored unfixed were positive by PCR.
This outbreak provided a unique opportunity to study the incubation period, symptom duration and spectrum of symptoms of E. bieneusi in otherwise healthy persons, which until now was largely unknown. For the first time, we have been able to document incubation periods for cases of microsporidiosis. However, because this was an observational study, it is important to consider the factors that could have influenced the wide range of incubation periods we report here. First, it is plausible that differences in dose response could account for some of the variation because it can be expected that those persons who consumed large amounts of the implicated foods might have had shorter incubation periods. Another factor which might explain the variation is differences in host immunity. Although we believe that our case-patients were immunocompetent persons, we did not systematically collect information about their medical history. We therefore performed a follow-up cohort study to investigate this possibility further (data will be published separately). The lack of specificity in the definition of several of the symptoms might explain some of the variation. Neither the first case-patient nor any of the case-patients with late-onset dates were microbiologically confirmed. The range of incubation periods for the four confirmed case-patients (3–15 days) is therefore more reliable and comparable to other protozoan parasites such as Cryptosporidium and Giardia: 1–12 days and 3–25 days, respectively [Reference Heymann29].
Our findings indicate that E. bieneusi is capable of causing an outbreak of gastrointestinal illness in a group of apparently immunocompetent persons. Whether E. bieneusi is newly emerging as a potential agent of foodborne outbreaks or whether it has simply been under-reported previously remains unclear but it is believed that foodborne protozoan infections are almost certainly under-detected by a factor of ⩾10 [Reference Casemore30]. Moreover, the aetiological agent remains unknown for more than half of all reported foodborne outbreaks in Sweden [Reference Lindblad31]. We hope that by reporting the findings of this investigation, the awareness of considering parasites such as microsporidia as possible agents during outbreaks will be raised, allowing for easier recognition of outbreaks. In particular, we recommend that microsporidia be explored as causative agents in food- and waterborne outbreaks involving long incubation periods (⩾1 week), predominantly non-bloody diarrhoea, abdominal pain and bloating, especially when no other organism has been identified.
ACKNOWLEDGEMENTS
We acknowledge Eva Andersson and Ingrid Persson for their help during the initial investigation and prevalence study. We thank Annika Hulu and Lotta Bertilsson for carrying out the environmental investigation and Yen Ngo and Anna-Maria Kling for statistical support. We thank Thomas Ahlqvist for performing the initial laboratory investigations and Henrik Vedel Nielsen for performing the real-time PCR. Finally, we thank Viviane Bremer and Katharina Alpers (EPIET) for reviewing this manuscript and Margarita Riera-Montes for support during the investigation. The EPIET fellowship of V. Decraene is funded by the European Commission DG SANCO.
DECLARATION OF INTEREST
None.






