Both dengue and chikungunya viruses have emerged as intriguing public health problems in tropical and subtropical areas in recent times [Reference Dash1]. The genome of both viruses is an enveloped, single-stranded, positive-sense RNA of ~12 kb. Dengue virus (DENV) infection generally causes a mild febrile illness, referred to as dengue fever (DF) but it may also lead to severe life-threatening dengue haemorrhagic fever (DHF) or dengue shock syndrome (DSS). In India, the first dengue outbreak was recorded in 1945 [Reference Sabin2]. Delhi experienced its first major DHF outbreak of DENV-2 genotype IV in 1996 followed by DF outbreaks of DENV-3 genotype III in 2003 and 2006 but in 2006 DENV-1 genotype III also co-circulated [Reference Dar3–Reference Kukreti5].
Chikungunya virus (CHIKV) was isolated first in Tanzania during a dengue outbreak in 1953 [Reference Ross6]. The presence of CHIKV is well documented in Asia and tropical Africa. In India, CHIKV outbreaks were documented at various locations during 1963–1973 [Reference Yergolkar7]. CHIKV re-emerged after a lengthy gap of 32 years in 2005–2006 with altered virulence in Indian Ocean Islands [Reference Schuffenecker8]. In 2006 a major outbreak was reported from Central and South India, although some sporadic confirmed cases were also reported from North India [Reference Chhabra9]. Co-infection cases of dengue and chikungunya viruses were also reported in 1967 and 2006 in Calcutta and Delhi, respectively [Reference Myers and Carey10, Reference Chahar11].
Aedes aegypti is the principal vector of both DENV and CHIKV in urban and semi-urban areas of India. Delhi has humid, monsoon-influenced subtropical weather and daily temperature ranging from 25°C to 30°C, which provide a conducive environment for the breeding of Aedes mosquitoes. The mosquito vector thrives well and transmits both DENV and CHIKV in the human population which may result in major outbreaks during the post-monsoon season. This led us to identify the aetiology of the 2010 outbreak by investigating a large number of patients that had characteristics of both types of viral infection from August to November 2010 in the Delhi region.
Acute-phase blood samples were collected from persons suspected as clinical cases of dengue and chikungunya fever who were referred to the National Centre for Disease Control (NCDC) from different locations in Delhi from August to November 2010. Patients who had constitutional symptoms like headache, joint pain, rash and fever of <5 days' duration were considered. These were outbreak-related samples, therefore no ethical approval was required.
The standard procedure was followed for isolation of viral RNA from serum samples (140 μl) using QIAamp Viral RNA Mini kit (Qiagen, Germany). Duplex reverse transcription–polymerase chain reaction (D-RT–PCR) was performed with the Access Quick one-step RT–PCR kit (Promega, USA) using primers for the CprM gene junction of DENV and the E1 gene region of CHIKV [Reference Dash1]. Reverse transcription was performed at 45°C for 30 min followed by PCR: initial denaturation at 95°C for 2 min, followed by 35 cycles of 94°C for 30 s, 55°C for 60 s, 72°C for 60 s and a final extension at 72°C for 10 min using an ABI 9700 Thermal cycler (Applied Biosystems, USA). The amplified fragments (511 bp for dengue, 205 bp for chikungunya and both for co-infection) were visualized after electrophoresis on ethidium bromide-stained 1·2% agarose gel. The PCR products were purified using QIAquick PCR purification kit (Qiagen, Germany). Sequencing was performed using the Big Dye Terminator cycle sequencing ready reaction kit v. 3.1 (Applied Biosystems, USA) on an automatic DNA sequencer (ABI 3130xl Genetic Analyzer, Applied Biosystems). Sequence alignment was performed using ClustalW multiple alignment. BLAST search was performed to confirm the virus type for dengue and chikungunya (www.ncbi.nlm.nih.gov/Education/BLASTinfo/information3.html). Sequences were submitted to Genbank and accession numbers acquired (Table 1). Phylogenetic analysis was performed using Molecular Evolutionary Genetics Analysis (MEGA) software version 4 and a phylogenetic tree was constructed using the maximum-likelihood (ML) method with a bootstrap value of 1000 replications.
Table 1. Details of patients sequenced in the studyFootnote *
Using D-RT–PCR, 93/432 serum samples were found positive for dengue and chikungunya which included five co-infection cases during August–November 2010 in Delhi. Fifty-four samples were diagnosed positive for DENV and 39 samples positive for CHIKV infection. Out of 93 samples 60 were male and 33 were female. Positive samples were related to different age groups, 12 samples were from the ⩾60 years age group, 75 samples from the 15–59 years age group and six samples from the <15 years age group. The most common clinical symptoms were fever (100%), headache (92%), myalgia (74·6%), and joint pain (68·3%). Twenty-two percent cases had a low thrombocyte count (<100 000/mm3). In co-infection cases a low thrombocyte count was reported in 3/5 samples.
To trace the type of circulating virus 34/54 D-RT–PCR positive samples of the CprM gene of DENV were sequenced and subjected to BLAST search which confirmed the circulation of DENV-1 genotype III. Similarly, 22/39 positive samples of the E1 gene of CHIKV were subjected to sequencing and all of them were found to be related to the East Central South African (ECSA) genotype. However, five co-infection cases also had the same genotype for both viruses. A phylogenetic tree was generated for DENV on the basis of the sequences of the present study and previously reported DENV-1 (Fig. 1). CprM gene sequences revealed clustering of isolates from different countries in three distinct genotypes (genotypes I, II, III). However, it is of note that all Indian DENV-1 sequences belonging to different outbreaks, clustered in genotype III in close proximity with the sequences from Comoros and Thailand.
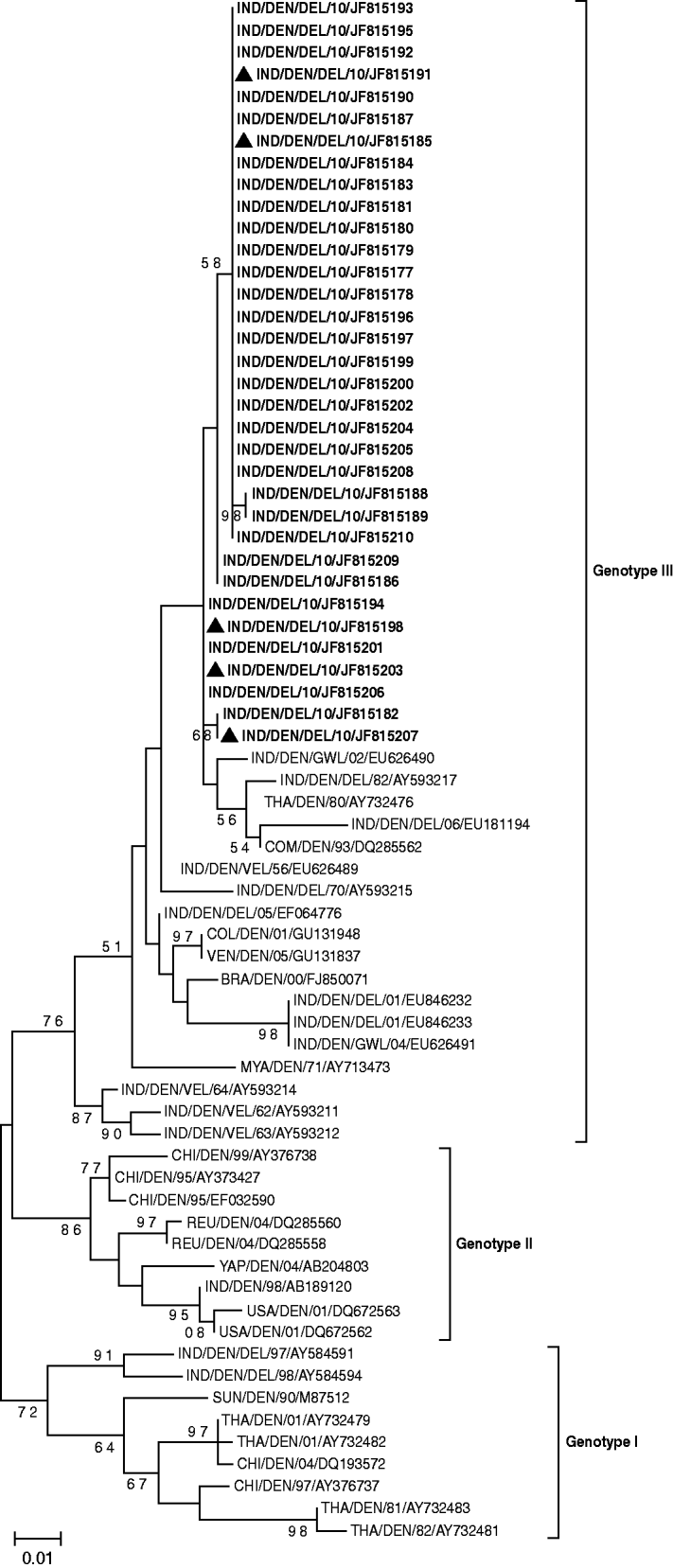
Fig. 1. Phylogenetic tree of DENV-1 based on 328-bp nucleotide sequences of the CprM gene region is generated by the maximum-likelihood method. Bootstrap support values (based on 1000 replications) above 50% are shown at the branch nodes. Each strain is denoted by country of origin, virus name, state name (only for Indian samples) followed by the last two digits of year of isolation and Genbank accession number. DENV-1 sequences that were sequenced in the study are shown in bold and co-infection cases are indicated by a triangle.
The phylogenetic tree constructed using the E1 gene studied CHIKV sequences and referred sequences from different parts of the world segregated into three genotypes (Fig. 2). Twenty-two of the CHIKV sequences of the present study clustered with the ECSA genotype along with previously reported Indian and global sequences from Indian Ocean Islands, S-27 African prototype, Tanzania, South Africa, Uganda and Republic of Congo. The earlier chikungunya Indian isolates (1963–1973) along with a large number of isolates from Thailand, Philippines and Indonesia formed a separate Asian genotype cluster. The sequences from Senegal and Nigeria clustered together to form the West African genotype.
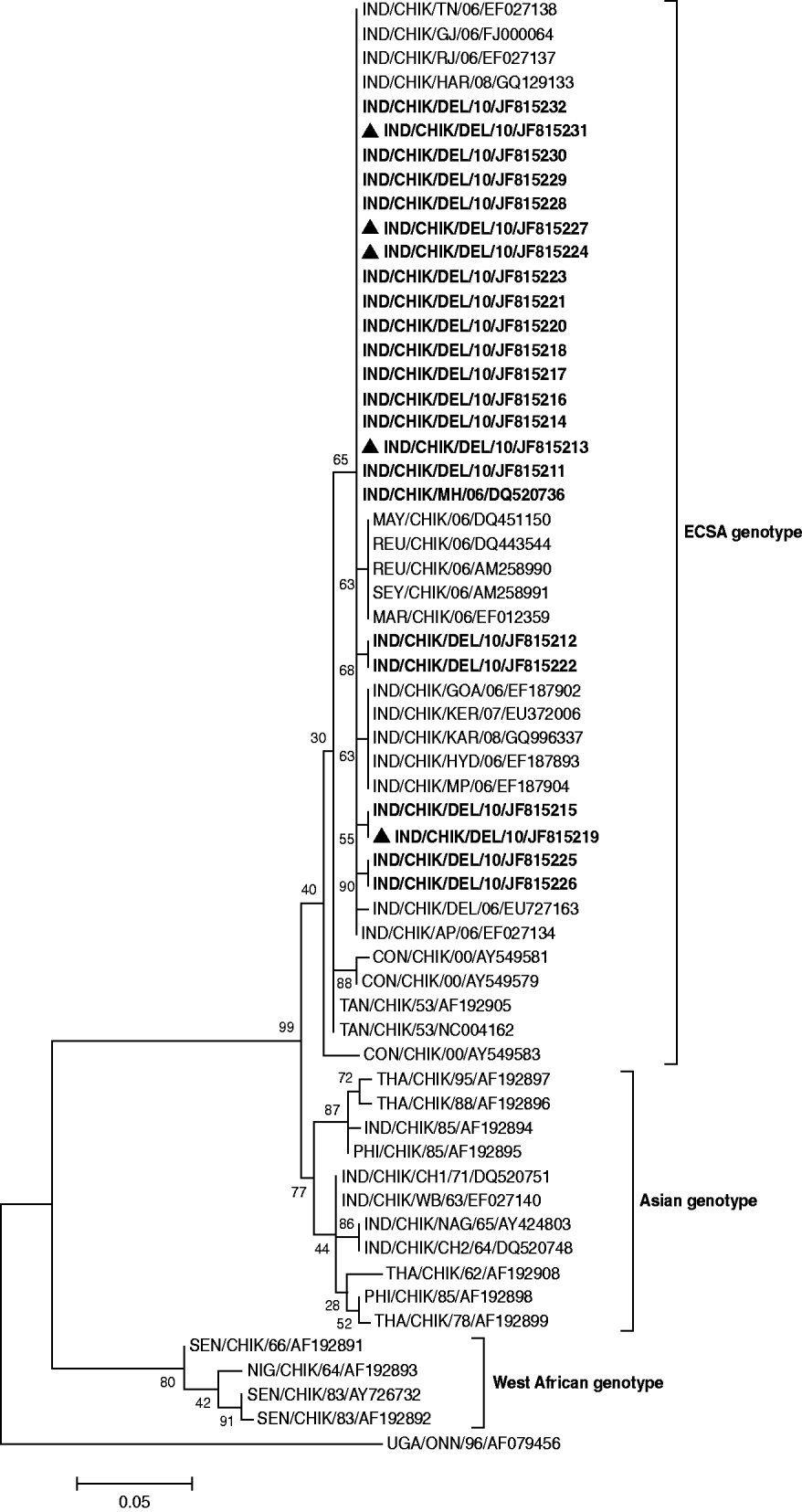
Fig. 2. Phylogenetic tree of CHIKV based on 205-bp nucleotide sequences of the E1 gene region is generated by the maximum-likelihood method. Bootstrap support values (based on 1000 replications) above 50% are shown at the branch nodes. Each strain is denoted by country of origin, virus name, state name (only for Indian samples) followed by the last two digits of year of isolation and Genbank accession number. O'nyong-nyong virus sequence was used as outgroup. CHIKV sequences that were sequenced in the study are shown in bold and co-infection cases are indicated by a triangle.
Most parts of the world have experienced rapid climate and demographic changes which lead to the emergence and re-emergence of Aedes species in hitherto unknown areas. Delhi is endemic for the co-circulation of multiple serotypes and genotypes of DENV which is known to cause periodic outbreaks [Reference Dar12]. The metropolitan city of Delhi experienced almost 33% of the total annual cases reported in India. In the Delhi 2006 dengue outbreak, some cases of chikungunya were also reported but in subsequent years (2007–2009) the number of cases of both viruses remained low [13]. Presently, no specific therapy and vaccine is available against both types of infection; hence, detection of the aetiology of an outbreak at the initial phase is very important. Circulation of CHIKV is seen in dengue endemic areas. The clinical manifestation of CHIKV infection mimics DENV infection in these areas, and it has been postulated that many CHIKV infections are misdiagnosed as DENV, thus causing CHIKV infections to be underreported [Reference Powers14]. During the 2010 outbreak, we used D-RT–PCR to find the probability of co-circulation of both viruses. An extensive increase in CHIKV cases compared to previous years along with DENV were recorded during the 2010 outbreak.
In conclusion, a dominant presence of CHIKV was found in the Delhi region during 2010, with a several-fold increase in the number of cases compared to those found in the 2006 DF outbreak. Interestingly, for the first time, we found that both DENV-I (genotype III) and CHIKV (ECSA genotype) co-circulated with equal dominance during August to November 2010 in Delhi. In addition, we also found cases of co-infection in circulation during the same period. The widespread emergence of DENV and exponential increase in CHIKV cases in Delhi during 2010 warrants the need for more effective surveillance to monitor the spread of these deadly arboviruses so that timely control strategies can be implemented.
ACKNOWLEDGEMENTS
The authors gratefully acknowledge the financial support from the National Centre for Disease Control (NCDC) during the course of this study.
DECLARATION OF INTEREST
None.





