Equine encephalosis (EE) is an arthropod-borne non-contagious viral infection, affecting all species of equids, usually characterized by mild or subclinical infection but occasionally causing severe clinical disease. The causal agent, equine encephalosis virus (EEV) is transmitted through the bites of certain Culicoides spp. EEV is also classified as a distinct virus species within the genus Orbivirus, family Reoviridae. Other species within the genus include African Horse Sickness virus (AHSV) and Bluetongue virus (BTV). All equids (horses, donkeys, zebras) are susceptible to EEV, although donkeys and zebras are thought to be resistant to the clinical disease.
EEV was first identified in South Africa in 1967 and is now considered endemic in South African equid populations. Studies in South Africa have reported evidence of exposure to EEV as high as 57–77% in horses and ponies, 49–85% in donkeys and 60–88% in zebras, as determined by seropositivity [Reference Hinchcliff, Sellon and Long1, Reference Howell, Guthrie, Coetzer, Coetzer and Tustin2]. Seven serotypes of EEV have been identified and horses can be simultaneously infected with multiple strains [Reference Howell3]. In a study performed on zebras in South Africa, neutralizing antibodies to all seven serotypes were identified [Reference Williams, Du Plessis and Van Wyngaardt4].
EEV infection is usually asymptomatic and the majority of cases are confirmed by serology. Clinical signs of disease, if observed, have only been recorded in horses and occur after an incubation period of 2–6 days. The signs are usually mild, but may vary from mild fever, listlessness and inappetance to, occasionally, depression, facial oedema, respiratory distress and cardiac signs [Reference Hinchcliff, Sellon and Long1, Reference Howell5]. The mortality rate associated with EEV infection appears to be very low.
In 2008/2009 a febrile horse disease, later diagnosed as EE, was observed for the first time in Israel in more than 60 equine premises across the country [Reference Mildenberg6]. Morbidity ranged from 2% to 100% and clinical signs in horses included raised body temperatures, oedema of the neck, legs, lips and eyelids, accelerated pulse/breathing rates and congested mucosa; however, none of the affected horses died from the disease. This was the first time that this disease had been observed outside southern Africa, which highlights the risks of its spread to other areas where competent vectors and susceptible animals are present, and emphasizes the need for improved knowledge and understanding of the extent of spread and circulation of this virus in countries beyond southern Africa. As mortality is considered to be very low and the virus is often thought to manifest itself subclinically, it is possible that EEV may be circulating undetected in countries beyond southern Africa, and the recent outbreak in Israel indicates that this may well be the case.
The aim of this study was to gain a better understanding of the distribution and levels of circulation of EEV beyond its known heartland in southern Africa. Blood samples (serum and, when possible, EDTA) were collected from equid populations in two West African countries (Ghana and The Gambia), one East African country (Ethiopia) and one North African country (Morocco). The results of the serological surveys of the equid populations for EEV antibodies, as well as the location of recent outbreaks of EEV in Africa, are illustrated in Figure 1. Antibodies to EEV were measured using a competitive ELISA (cELISA) previously shown to have a sensitivity and specificity of 100% [Reference Crafford7]. The presence of EEV RNA was detected using a real-time RT–PCR assay. This assay is currently being commercialized with Laboratoire Service International (LSI) and a paper describing the assay is in preparation.
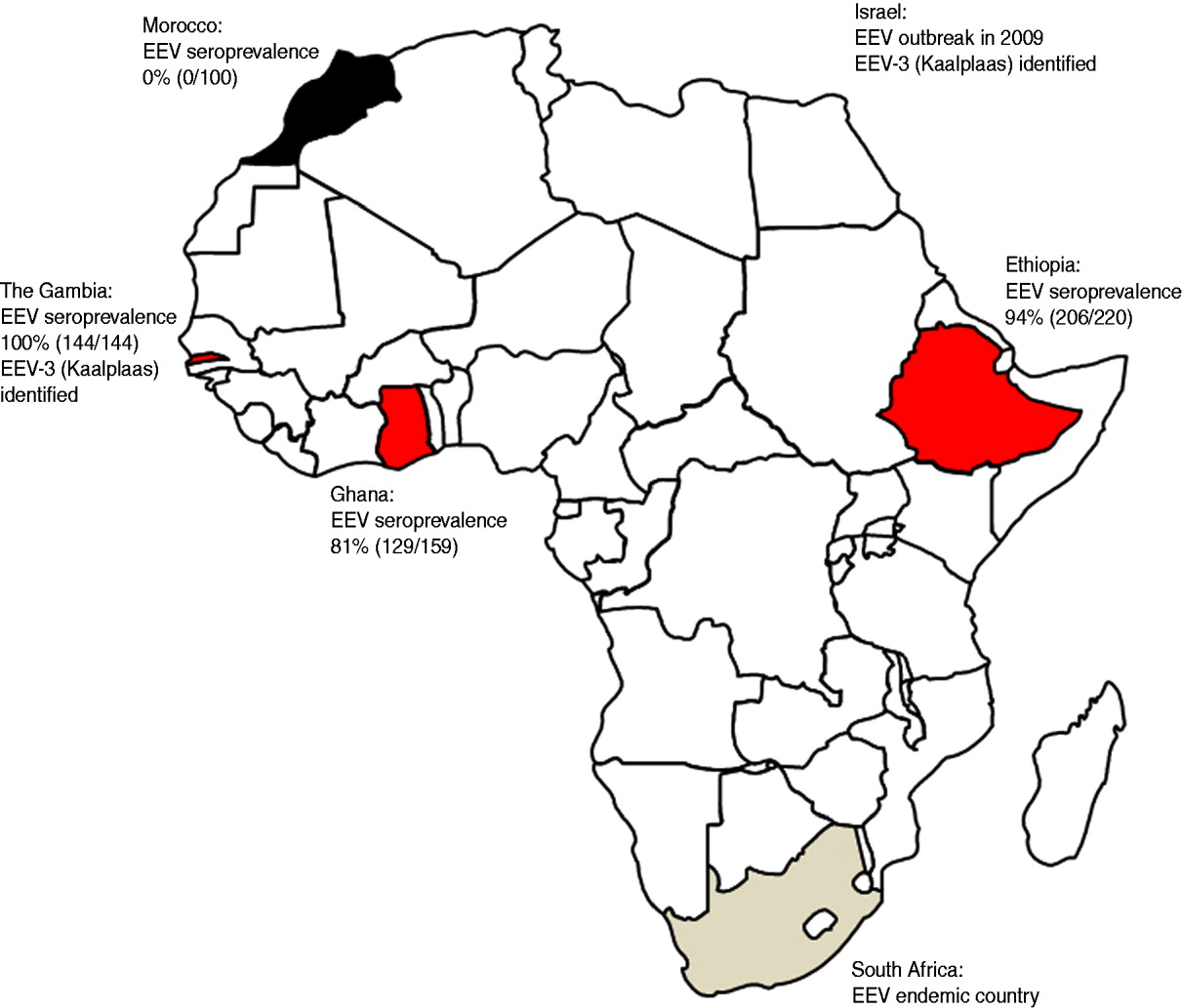
Fig. 1. Map of Africa showing the seroprevalence of antibodies to equine encephalosis virus (EEV) in equid populations from Morocco, The Gambia, Ghana and Ethiopia. The location of recent outbreaks of equine encephalosis (EE) in both South Africa and Israel are shown for interest. EEV-3, Equine encephalosis virus serotype 3.
The Gambia: In October 2009 a serological and virological survey for EEV and AHS was undertaken. In total 144 equids (horses and donkeys) were sampled from seven villages in the north Nianija district in mid-Gambia and 10 villages south of the river Gambia in the district of Niamina. Blood samples (serum and EDTA) were tested for antibodies to EEV by ELISA and for EEV RNA by real time RT–PCR. The median age of the equids (101 horses, 43 donkeys) was 7 years (range 1–20 years). All of the 144 equids sampled, including samples taken from equids as young as 18 months old, were seropositive for EEV antibodies as determined by ELISA [Reference Crafford7]. The equids had no previous history of showing clinical signs associated with EE; however, it is possible that clinical signs may have been missed due to the extensive management systems conducted in The Gambia. These serological and clinical data indicate that EEV is circulating freely in the region and is infecting equids at an early age, but there is no evidence that the virus is causing significant clinical disease in the infected animals.
Out of the 144 sampled equids, six young donkeys that were aged <3 years, tested positive for EEV RNA by real-time RT–PCR assay. EEV was isolated directly on BHK cells from two of these samples (IAH reference collection nos. GAM2009/05 and GAM2009/06) and the two isolated samples tested positive by conventional RT–PCR using primers targeting Seg-10 [Reference Aharonson-Raz8]. Full-length cDNA copies of individual EEV genome segments were synthesized and amplified from GAM2009/05 by RT–PCR, using the ‘anchor spacer–ligation’ method as described previously [Reference Maan9, Reference Potgieter10]. Partial sequences (for the upstream 450 bp; GenBank accession no. JN391443) of Seg-2, showed 92·8% nucleotide (nt) and 96·4% amino acid (aa) sequence identity with Seg-2 and VP2 of the ‘Kaalplaas’ reference isolate of equine encephalosis virus serotype 3 (EEV-3) (accession no. HQ630933) (Fig. 2). An EEV strain, isolated from horses during a recent outbreak in Israel, was also identified as EEV serotype 3 [Reference Aharonson-Raz8]. Subsequent sequence analysis of Seg-2/VP2 from the Israeli isolates (IAH reference collection nos. ISR2009/20 and ISR2009/21, accession nos. JF495411 and JF495412, respectively) and The Gambian isolates showed a very high level of sequence identity (98·8% nt/99·3% aa) indicating that they both belong to the same serotype, and were recently derived from a common ancestor (Fig. 2). Based on previous phylogenetic comparisons, different BTV serotypes, Seg-2/VP2 showed a maximum of 71% nt and 78% aa sequence identity between serotypes [Reference Maan9]. This indicates that The Gambian isolate, which (like the Israeli isolate) shows >92% nt and >96% aa identity to other EEV-3 strains in Seg-2/VP2 also belong to EEV serotype 3.
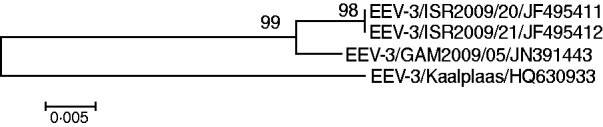
Fig. 2. Neighbour-joining tree, showing relationships in Seg-2 (upstream 450 bp) between the reference strain of equine encephalosis virus serotype 3 (EEV-3/Kaalplaas) and field strains of EEV-3 from The Gambia (EEV-3/GAM2009) and Israel (EEV-3/ISR2009). The tree was constructed using distance matrices, generated using the P distance determination algorithm in MEGA 4.1 (500 bootstrap replicates) [Reference Tamura12]. Scale represents number of substitutions per site. Values at major branching points represent NJ bootstraps.
Ghana: In August 2010 serum samples were collected from adult horses in the Accra region of Ghana. EDTA blood samples were not collected. Most of the horses were born in Ghana with a few being brought in from Nigeria and South Africa. The horses were used in polo games and regularly travelled between Ghana and Nigeria for tournaments. Out of the 159 serum samples collected 129 (81%) tested positive for EEV antibodies as determined by ELISA [Reference Crafford7] indicating that EEV is circulating freely in the region and infecting equids. The equids had no previous history of showing clinical signs associated with EE and, as the horses were generally well managed, it is likely that clinical signs would have been noticed if they had occurred.
Ethiopia: In 2008 a total of 220 sera and EDTA blood samples were collected from AHS suspected outbreak sites across Ethiopia. Samples were taken from 72 adult horses from Jijiga town in the Somali region of Eastern Ethiopia, 135 horses, donkeys and mules of mixed ages (mostly adult) from the South West of Ethiopia and 13 adult horses from the Bale region of South East Ethiopia.
All EDTA blood samples tested negative for EEV RNA by real-time RT–PCR. However, 206 out of the 220 (94%) serum samples tested positive for EEV antibodies as determined by ELISA [Reference Crafford7], indicating that EEV is circulating freely in the region and infecting equids from a young age. Due to the extensive management systems of these equids in Ethiopia it was not clear if the animals had previously shown clinical signs of EE.
Morocco: In November and December 2007, 120 serum and EDTA blood samples were collected from mixed-age groups of horses on two farms in Morocco (Marrakech in central Morocco and Larache in northern Morocco). All 120 EDTA-treated blood and serum samples were negative for both EEV RNA and antibodies when tested by RT–PCR and ELISA, respectively. This indicated that EEV was not circulating in the equine populations within these regions of Morocco.
EE has long been known to be enzootic in southern Africa, but up to now has not been reported to have been detected either virologically or serologically in eastern/central/western or North African countries. We report a high prevalence of EEV antibodies in equids from countries in western and eastern Africa. We conclude that EEV is likely to be endemic in equids in The Gambia, Ghana and Ethiopia, with horses and donkeys in the region being infected with the virus from a young age. The circulating serotype in The Gambia was identified as EEV-3 (Kaalplaas) which is the same serotype identified during the recent outbreak of EE in Israel [Reference Mildenberg6]. Additionally sequence comparison of Seg-2/VP2 genes from the Israeli and The Gambian isolates revealed a very high level of sequence identity (98·8% nt/99·3% aa). The close genetic relationship between these two isolates indicates that they are likely to have been derived from a common ancestor. This emphasizes the importance of conducting further surveys in other countries of central, eastern and northern Africa in order to identify the levels of circulation and to determine the infection routes for EEV. Interestingly, horses in Morocco tested negative for EEV antibodies indicating that the Sahara desert may act as a geographical barrier to the spread of infected insects and therefore the virus to North African countries.
The majority of information about the clinical signs and severity of EEV infections is from South Africa where the disease has been considered endemic for many years. In this endemic environment more than 90% of animals show either no obvious clinical signs of infection, or they develop only mild clinical signs. Occasionally, more severe clinical signs are seen in horses from which EEV has been isolated [Reference Erasmus, Boshoff, Pieterse, Bryans and Gerber11]; however, it is not always clear whether EEV was the primary cause of disease. The recent outbreak of EEV in Israel, however, has shown that the virus is capable of causing severe outbreaks of disease when circulating in a naive population. The route of spread of EEV to Israel remains unknown; however, it is possible that it spread either directly via Egypt, or through the horn of Africa via Middle Eastern countries. Further studies are required in Middle Eastern and North African countries in order to gain a fuller understanding of where EEV is circulating and the associated risks of its transmission and spread. Further work is also needed to investigate the clinical significance of EEV infection in naive populations of athletic/thoroughbred horses.
This study shows, for the first time, that EEV is endemic and freely circulating in equid populations in countries in East and West Africa; however, there was no evidence of EEV circulation north of the Sahara desert in Morocco. As EEV is an arbovirus that is transmitted by Culicoides midges, there is a high risk of further spread once the virus is introduced into a new area where Culicoides midges are present. It therefore becomes important to know where this virus is circulating so that appropriate control measures can be put in place to stop further spread to EEV-free countries. Currently, as EE is not a notifiable disease, it is not necessary to test animals prior to movement, which increases the risk that this virus may be transported to disease-free countries through the movement of infected equids.
ACKNOWLEDGEMENTS
We thank the Department of Veterinary Services, The Gambia, and in particular Mr Borrie Jabang, Laboratory Technician at the Central Veterinary Laboratory, Abuko. We thank the entire local staff of The Gambia Horse and Donkey Trust, Sambel Kunda, and animal owners for providing their animals for sampling. We also thank the BEVA Trust for funding the field work in The Gambia. We also thank The Polo Club in Accra, Ghana for allowing their horses to be sampled and both Abid BinTarif and Kyriaki Nomikou for helping to process the samples at IAH, Pirbright. We also acknowledge financial support from Defra and BBSRC.
DECLARATION OF INTEREST
None.




