INTRODUCTION
West Nile virus (WNV) is a mosquito-borne arbovirus belonging to the genus Flavivirus (family Flaviviridae). Main reservoir hosts are birds but the virus also infects various species including horses and humans, with marked consequences for public health and for the equine industry due to potentially fatal encephalitis [Reference Campbell1, Reference Castillo-Olivares and Wood2]. Since the discovery of WNV in 1937 in Uganda [Reference Smithburn3], the geographical distribution of the virus has expanded widely [Reference Campbell1, Reference Ozdenerol, Taff and Akkus4]. In Europe, WNV was first recognized in 1962 in France. Several outbreaks have since been documented in many European countries [Reference Calistri5], and increasingly so in southern and eastern Europe (e.g. Italy, Greece, Bulgaria, Croatia, Serbia, Albania) [Reference Di Sabatino6], resulting in the virus now being considered endemic in large parts of Europe.
The recent introduction of Lineage 2 in Europe [Reference Bakonyi7–Reference Hernández-Triana9] resulted in more severe clinical cases in humans [Reference Danis10] and contributed to WNV becoming a growing public health concern in Europe in general, and in France in particular. French outbreaks occurred between 2000 and 2006 when a total of 114 confirmed equine cases and four confirmed human cases were reported [Reference Del Giudice11–14]. In summer 2015, new WNV-confirmed cases were reported in southern France in 49 horses and one human [Reference Bahuon15]. In most countries, including France, the surveillance of WNV is mainly passive (i.e. examination of clinically affected cases of specified diseases only in the population). However, the performance of passive surveillance systems suffers from frequent under-reporting especially for the surveillance of exotic diseases which have a low probability of occurrence [Reference Doherr and Audigé16]. This may result in a failure to identify the disease and thus, the main challenge in improving early detection of WNV outbreaks is the development of more efficient early detection systems for limiting the consequences of a WNV outbreak in both equine and human populations.
Syndromic surveillance is defined as the (near) real-time collection, analysis, interpretation and dissemination of non-specific health-related data to enable the early identification of potential threats like a disease [17]. Nervous syndromes in horses are considered to be an early indicator of WNV outbreaks [Reference Leblond, Hendrikx and Sabatier18, Reference Saegerman19] and, using them in a syndromic surveillance system, might be one of the most cost-effective surveillance systems in the European context [Reference Chevalier, Lecollinet and Durand20]. In France, the passive French surveillance system RESPE [Réseau d'Epidémio-Surveillance en Pathologie Equine; the French network for the surveillance of equine diseases (http://www.respe.net/)], collects declarations from veterinary practitioners registered as sentinels throughout France. Data on nervous signs observed in French horses have been collected since 2006. More than 550 sentinel veterinarians are involved, providing coverage of 92 out of 96 French regions. The veterinarians complete a standardized questionnaire online and send standardized samples for laboratory diagnosis. Diagnostic tests for WNV, equine herpes virus serotype 1 (EHV-1), and other types of herpes viruses (EHV-sp) [Reference Léon21] are systematically implemented for each declaration of nervous signs. Using routinely collected RESPE data in an early detection surveillance system could lead to the timelier implementation of protective measures before laboratory test results. Currently, the collected RESPE data on nervous signs are mainly used to produce alerts when cases with positive laboratory diagnoses are identified. The data are also used for basic syndromic surveillance: an alarm is triggered when four declarations are reported in the same week, or three declarations reported each week for two consecutive weeks. This alarm threshold was set arbitrarily and alarms may result in the initiation of epidemiological investigations depending on the context of the declarations. However, the reliability of this threshold has never been assessed and the ability of the RESPE nervous syndrome database to serve as a routine syndromic surveillance system is currently unknown.
Our objective in this study was to assess whether or not RESPE data can be used as a routine syndromic surveillance system for the detection of WNV outbreaks in France testing several pre-processing methods and detection algorithms to model time-series data. We evaluated system performances using simulated and authentic data and compared them to those of the surveillance system currently in place.
METHODS
Data characterization
In the RESPE database, nervous signs in horses are defined as any signs of impairment of the central nervous system, i.e. ataxia, paresis, paralysis and/or recumbency, and/or behavioural disorder. Cases, or an unusual cluster of cases, with ‘atypical’ expression (colic, lameness, excitement, falling, muscular atrophy) can also be considered after the most common aetiologies for these signs have been excluded. These signs can indeed be the clinical manifestation of a central nervous system disease. Nervous disorders with evidence of traumatic or congenital origins are excluded.
Data on nervous signs in horses were available from RESPE for every calendar day from 1 January 2006 to 16 October 2015, totalling 653 declarations. However, in the remainder of the study, the time series was aggregated into weekly counts due to the low per-day count. Monthly aggregation was not considered, as the main objective of this surveillance system was early detection.
Tests for WNV and EHV are routinely carried out on horses that present nervous signs, and the database contains positive laboratory results mainly for EHV-1 but also some WNV cases. The EHV-1-positive cases were either isolated cases, i.e. not associated with other positive cases, or from a cluster of cases that could represent a true outbreak.
Models
We split the data into three sets:
-
• Set 1. Data from 2006 to 2010 were used to train our models.
-
• Set 2. Data from 2011 to 2013 were used to validate our models, simulate nervous signs time series and evaluate detection performances using simulated time series and outbreaks.
-
• Set 3. Finally, raw data from 2014 and 2015 were used to evaluate detection performances using real data as if a syndromic surveillance system was implemented on the field based on models estimated using sets 1 and 2. WNV outbreak occurring in France in autumn 2015 [Reference Bahuon15] was used to test our system with a real outbreak.
Data cleaning (set 1)
The raw time series from 2006 to 2010 was designated TS0. We investigated three methods for the removal of aberrations present in TS0 in order to obtain an outbreak-free baseline. In the first method, we retained only the 452 cases with no positive laboratory results (TS1). The second method consisted of removing all data linked to historical EHV-1 outbreaks, based on information from the RESPE website (TS2). This method did not remove single positive cases but only the positive cases associated with a cluster of other positive cases. In our third method, extreme values from TS0 were removed using the approach of Tsui and colleagues [Reference Tsui22], which assumes that, after the data have been fitted to a regression model, data points above the 95% confidence interval of the model prediction represent an outbreak (TS3). The authors used Serfling's regression model [Reference Serfling23], which is a linear regression model that uses sine and cosine terms to account for seasonal variation. With our own data, we followed the proposal of Dórea and colleagues [Reference Dórea24] and used a Poisson regression, which they considered an appropriate method to capture baseline activity while minimizing the influence of aberrations present in the dataset. The data were thus first fitted to a Poisson distribution (see model in the Supplementary material) and then values above the 95% confidence interval were removed. In TS1, TS2, and TS3, the values of the weeks considered to be part of an outbreak were not just removed but instead replaced by the average of the four previous weeks. The four time series are shown in Figure 1.

Fig. 1. Four time series used. TS0, raw data; TS1, only the cases with no positive laboratory results for WNV or EHV-1; TS2, outbreaks removed based on historical data; TS3, extreme values above the 95% confidence interval deleted.
The explainable patterns (such as global linear trends and seasonality) were investigated in each time series (TS0, TS1, TS2, TS3) in order to assess the impact of pre-processing methods on the dataset. We generated summary statistics by month and year, and performed moving average and autocorrelogram analysis [Reference Lotze, Murphy and Shmueli25].
Model training (set 1) and validation (set 2)
Modelling was attempted using generalized linear regression models (GLMs) that were appropriate for count data [Poisson and negative binomial (NB) regressions] and Holt–Winters generalized exponential smoothing (HW). For GLMs the evaluated models included different types of seasonality through the use of sinusoid functions with one, two, or three periods/year and season or month as factorial variables. To account for differences between years, we also calculated the average counts over 53 consecutive weeks (histmean). To ensure that an ongoing outbreak would not influence the estimate, we used a 10-week guard band for the calculation of histmean. A list of tested variables is available in the Supplementary material.
Alternative GLMs were evaluated on the training data from 2006 to 2010 using Akaike's information criterion (AIC) [Reference Bozdogan26]. For the HW method, the optimal parameters were determined through minimization of the squared prediction error [Reference Kalekar27]. The best models were then evaluated and compared using the autocorrelation and partial autocorrelation functions of the residuals (ACF and PACF, respectively) and the root-mean-squared error (RMSE). ACF is the linear dependence of a variable on itself at two points in time and PACF is the autocorrelation between two points in time after removing any linear dependence between them [Reference Box, Jenkins and Reinsel28]. ACF and PACF are used to find repeating patterns (e.g. seasons) in a dataset. RMSE is a measure of the difference between the values predicted by a model and the values actually observed from the environment that is being modelled [Reference Chai and Draxler29]. This criterion was calculated for the differences between the observations and the predicted values within both the calibration period (RMSEc) from 2006 to 2010 and the validation period (RMSEv) from 2011 to 2013. In either case, the lower the criterion, the better the predictive performance of the model.
Outbreaks detection
Simulated baselines and outbreaks (set 2)
We simulated 500 sets over 3 years. The model previously fitted to the raw historical baseline TS0 was used to predict value for each week of 2011, 2012 and 2013. Then, for each of our simulated years, the weekly number of nervous cases was randomly sampled from a Poisson distribution with a mean defined by the previous predicted value.
We simulated 500 WNV outbreaks based on historical data from five previous European outbreaks: French outbreaks in 2000 [Reference Murgue13], 2004 [Reference Leblond, Hendrikx and Sabatier18], 2006 [14], an Italian outbreak in 1998 [Reference Autorino30], and a Hungarian outbreak in 2008 [Reference Kutasi31]. The French outbreak in 2015 was preserved to evaluate detection performances using real data (set 3).The mean weekly count of nervous-symptom cases in horses was calculated from the five historical outbreaks for an outbreak period covering a total of 12 weeks, from the first positive case detected to the last positive case detected (see Fig. 2). The weekly number of WNV cases of a simulated outbreak was randomly sampled from a Poisson distribution with a mean defined by the previous mean weekly count.

Fig. 2. European West Nile virus outbreaks and nervous signs in horses. Number of confirmed cases per week between the first detected case and the last detected case. Dashed grey line indicates outbreak in Italy, 1998 [Reference Autorino30], dotted black line indicates outbreak in France, 2004 [Reference Leblond, Hendrikx and Sabatier18], solid black line indicates outbreak in France, 2000 [Reference Murgue13], dashed black line indicates outbreak in France, 2006 [14], solid grey line indicates Hungarian outbreak, 2008 [Reference Kutasi31]
Simulated outbreaks were randomly inserted in each set of 3 simulated years (see examples in Fig. 3). A total of 1500 years containing a total of 500 outbreaks were evaluated.

Fig. 3. Four examples of simulated data between 2011 and 2013 with one simulated outbreak inserted in each simulated dataset. Outbreak time periods are identified by dotted lines.
Authentic baseline and outbreak (set 3)
Raw data from 2014 and 2015 were used for the assessment of algorithm performances using authentic baseline and WNV outbreak going from week 33 to 44 (Fig. 4). Data from 2006 to 2013 were used to predict value for each week of 2014 and 2015. The three methods for the removal of aberrations previously tested were also applied to 2011, 2012 and 2013 in order to obtain complete outbreak-free baselines. These four new baselines from 2006 to 2013 were named TS0′, TS1′, TS2′ and TS3′ according to the method used in the previous section for the removal (or not) of aberrations.

Fig. 4. Raw data from 2014 to 2015 with West Nile virus outbreak identified with dotted lines.
Regarding WNV outbreak in autumn 2015 [Reference Bahuon15], only cases collected by RESPE were considered. Performances of our detection algorithm for this specific outbreak were assessed using the best alarm threshold previously identified with simulated data.
Detection algorithm
All eight combinations of pre-processing and forecasting methods were evaluated on their ability to detect disease outbreaks: GLMs applied to TS0, TS1, TS2, and TS3; and HW applied to TS0, TS1, TS2, and TS3. The outbreak detection method used was based on a multiple of the upper limit of the confidence interval of the prediction based on Serfling's approach [Reference Serfling23]. The alarm threshold was thus defined as the predicted number of cases in a given week plus a multiple of the standard error of the model prediction. If the actual observed value was above the threshold, an alarm was triggered. A 6-week guard band was used to ensure that detection algorithm was not disturbed by the first weeks of outbreak signal.
The detection performances of the surveillance system currently in place were also evaluated using as detection algorithm the RESPE's protocol: an alarm is triggered when four declarations are reported in the same week, or when three declarations are reported in each of two consecutive weeks.
Quantitative assessment
We first calculated sensitivity based on the number of outbreaks detected out of all inserted outbreaks and denoted this Se_out. An outbreak was detected when it triggered at least one true alarm, defined as a week that produced an alarm and that was a part of an epidemic period. Se_out was calculated as:
where Out is the number of outbreaks detected and No_Out is the number of outbreaks not detected.
We also calculated Se_wk, the sensitivity based on the number of weeks in an epidemic period in which an alarm was triggered. Se_wk and specificity (Sp) were calculated as:
where TP is the number of true positive alarms, TN the number of true negative alarms, FP the number of false-positive alarms, and FN the number of false-negative alarms.
A receiver-operating characteristic (ROC) curve was generated in R by testing various alarm thresholds, and the area under each curve (AUC) was also calculated [Reference Hanley and McNeil32]. The time to the first true alarm within an epidemic period was also evaluated in order to assess the efficiency of early detection.
Implementation
Models were implemented in R x64 version 3.0·2 [33]. Dynamic regression was performed with the functions glm (package ‘stats’), glm.nb (package ‘MASS’ [Reference Ripley34]), and stlf (package ‘forecast’ [Reference Hyndman35]). The expected numbers of counts at time t were estimated with the predict functions of the respective packages. The expected numbers of outbreak-related cases were estimated with the fitdist function of the package ‘fitdistrplus’ [Reference Delignette-Muller36]. AUCs were estimated with the auc function of the package ‘flux’ [Reference Jurasinski37].
RESULTS
Baseline characterization
At the week level, all baselines showed a significant increasing number of declarations. However, this trend was mainly due to the first years of data collection. A significant seasonal effect was also present in all time series: the number of declarations appeared highest in November, December, and January compared to other months. However, this seasonality was weak and principally apparent in the raw TS0 data, due to EHV-1 and EHV-sp outbreaks present in the dataset during the winters (see Supplementary material for details).
Assessment of models
The data from 2006 to 2010 for each time series were fitted to their respective appropriate regression model, using variables that accounted for seasonal effects. For the Poisson as well as the NB regression, the best fit was obtained for all time series with the simple model:
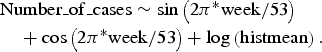 $$\eqalign{& {\rm Number}\_{\rm of}\_{\rm cases} \sim \sin \left(2{\rm \pi}^{\ast}{\rm week}/53 \right) \cr & \quad + \cos \left(2{\rm \pi}^{\ast} {\rm week}/53 \right) + \log \left({\rm histmean} \right).}$$
$$\eqalign{& {\rm Number}\_{\rm of}\_{\rm cases} \sim \sin \left(2{\rm \pi}^{\ast}{\rm week}/53 \right) \cr & \quad + \cos \left(2{\rm \pi}^{\ast} {\rm week}/53 \right) + \log \left({\rm histmean} \right).}$$
NB and Poisson regressions performed equally well for all time series, with the exception of TS0 (raw data), for which the NB model provided a better fit (AIC 749 vs. 761).
The details of differences between the smoothing performance of the best GLMs obtained and HW are presented in the Supplementary material. In all regression methods used, TS0 produced the worst results, while TS1 generated the best-fitting parameters. TS2 and TS3 yielded intermediary results, with better performances for TS3 than for TS2.
Outbreak detection
Simulated data
The results show that the AUCs of all methods and time series are small when the sensitivity used is based on the number of weeks within an epidemic period that produced an alarm (Se_wk) (see Table 1). This is consistent with the fact that first and last weeks within an epidemic period have very few cases which are difficult to detect. Using Se_wk, the ROC curves are similar between the different time series but the GLM always outperformed the HW method (see Fig. 5 and Table 1). By using instead the percentage of outbreaks detected (with at least one alarm) among all the outbreaks inserted (Se_out), the AUCs for all combinations of time series and methods improved to 0·98. The AUCs are similar for each pre-processing and forecasting method but the ROC, activity monitoring operation curve (AMOC) and free-response ROC curve (FROC) curves show differences (see Figs 6–8). The HW method outperformed the GLM in terms of detection performance using TS1 (i.e. better balance between sensitivity and specificity, and better balance between percentage of outbreaks detected and average number of false-positive alarms per year), and the GLM outperformed the HW method using TS0, TS2 and TS3 (i.e. better balance between sensitivity and specificity, and better balance between timeliness and average number of false-positive alarms per year).

Fig. 5. Receiver-operating characteristic (ROC) curves for each pre-processing and forecasting method representing median Se_wk (sensitivity based on the number of weeks within an epidemic period detected), plotted against median specificity, Sp. Error bars show the 25% and 75% percentile of the point value over 1500 simulated years and 500 simulated outbreaks. Blue point shows RESPE's current performances. GLM, Generalized linear model; HW, Holt–Winters.

Fig. 6. Receiver-operating characteristic (ROC) curves for each pre-processing and forecasting method representing median Se_out (sensitivity based on the number of outbreaks detected out of all inserted outbreaks), plotted against median specificity, Sp. Error bars show the 25% and 75% percentile of the point value over 1500 simulated years and 500 simulated outbreaks. HW, Holt–Winters; GLM, generalized linear model.

Fig. 7. Activity monitoring operation curves for each pre-processing and forecasting method representing median time for outbreak detection, plotted against number of false-positive alarms per year. Error bars show the 25% and 75% percentile of the point value over 1500 simulated years and 500 simulated outbreaks. Blue point indicates RESPE's current performance. HW, Holt–Winters; GLM, generalized linear model.

Fig. 8. Free-response ROC curves for each pre-processing and forecasting method representing the percentage of outbreaks detected, plotted against the number of false-positive alarms per year. Error bars show the 25% and 75% percentile of the point value over 1500 simulated years and 500 simulated outbreaks. Blue point indicates RESPE's current performance. HW, Holt–Winters; GLM, generalized linear model.
Table 1. Median value, 25th and 95th percentiles of the AUC (area under the receiver-operating characteristic curve) estimated for each pre-processing and forecasting method using Se_wk or Se_out

Se_wk, sensitivity based on the detection of every week which is a part of an epidemic period, Se_out, sensitivity based on the number of outbreaks detected out of all inserted outbreaks; GLM, generalized linear model; HW, Holt–Winters.
With the HW approach, the optimal balance between Se_out and specificity (Sp) was obtained when the alarm threshold equaled the upper limit of the confidence interval of the standard error of the model prediction multiplied by a constant close to 1·7 (Fig. 9). This alarm threshold detected more than 99% of the inserted outbreaks with an average time-to-detection of <3 weeks, and it produced between two and four false-positive alarms per year. The associated specificity was >0·96. Alarm thresholds based on constants higher than 1·7 had a lower Se_out (around 80% of outbreaks detected) and needed more time to produce the first true alarm (>3 weeks). With the GLM, the optimal balance between Se_out and Sp was obtained when the alarm threshold equalled the upper limit of the confidence interval of the standard error of the model prediction multiplied by a constant between 2 and 2·5 (see Fig. 9). Exact value varies according to the time series considered but, to detect at least 98% of the inserted outbreaks with an average time-to-detection close to 3 weeks, the associated specificity varied between 0·94 and 0·97 for a constant varying between 2·15 and 2·6 and a number of false-positive alarms between two and four per year.

Fig. 9. Curves for each pre-processing and forecasting method representing the Se_out (sensitivity based on the number of outbreaks detected out of all inserted outbreaks), and specificity, Sp, plotted against the alarm threshold k used for outbreak detection. GLM, Generalized linear model; HW, Holt–Winters.
Median performances obtained using the current RESPE alarm threshold were: Se_out equalled 99%, Sp near 90%, an average timeliness of 2·14 weeks, and between four and five false-positive alarms per year. These detection performances were consistent with those obtained with our models (see Figs 5, 7 and 8) and even outperformed them regarding Se_wk which reached 53% for an associated Sp of 90% when our regression models obtained a Se_wk below 50%.
Authentic data
To test our systems on real data, we used as a constant the value previously identified as providing optimal balance between Se_out and Sp, i.e. 1·7 for HW, and 2 for GLMs. Using these constants, all methods tested were able to detect the WNV outbreak. All time series using GLMs gave an alarm 4 weeks after the 2015 WNV outbreak started (week no. 37), had a specificity between 0·86 and 0·90 and produced more than four false alarm per year. Using the HW method, all time series gave an alarm 6 weeks after the outbreak started except TS1′ which was able to trigger the first alarm after 4 weeks. Their specificity ranged from 0·93 to 0·95 and their number of false-positive alarms was between two and three per year. The weekly sensitivity, Se_wk, was low for all methods tested and ranged from 0·12, for HW associated with TS0′ and TS2′, to 0·37 for TS1′ using HW and GLMs.
The alarm threshold currently used by RESPE also provided its first alarm on week 37 (i.e. 4 weeks after the start of the WNV outbreak). Its weekly sensitivity equalled 0·37 for a specificity of 0·95 and two false-positive alarms raised per year.
DISCUSSION
Our study shows that the RESPE data on nervous signs in horses could be used as an alarm system for WNV outbreaks in France. Regression models (i.e. GLMs or HW) and current RESPE alarm thresholds were able to detect WNV outbreaks and they performed similarly when we considered for regression models an alarm threshold defined to obtain the best balance between sensitivity, specificity and timeliness. The results obtained with simulated data indicated that such surveillance systems could detect >98% of WNV outbreaks with a specificity >94%, a timeliness between 2 and 3 weeks and an average number of four false alarms per year. According to our results, the alarm threshold currently used by RESPE is thus probably the best threshold that system managers could have found using a fixed alarm value throughout the year. These results are encouraging but this timeliness corresponds to the time needed to obtain a laboratory confirmation after clinical suspicion [Reference Bahuon15, Reference Leblond, Hendrikx and Sabatier18]. Using such alarm thresholds would thus only be of interest if WNV laboratory tests are not systematically implemented, which is currently not the case within RESPE. A better timeliness could be reached if the alarm threshold was modified. However, using a fixed value throughout the year as an alarm threshold does not take into account the seasonal variation in the number of cases reported. Regression approaches are able to deal with data seasonality and trend and they are thus more flexible and adaptable. Considering the specific situation of RESPE, even if regression models are more complex to implement than a fixed alarm threshold, they are also more interesting when the surveillance priority is not to reach the optimal balance between sensitivity, specificity and timeliness (e.g. surveillance priority could be to obtain better timeliness even if the number of false alarms increases).
Our study reveals differences in the time series and smoothing methods tested. As expected, the pre-processing methods that were used to remove past outbreaks present in the dataset modified the seasonality of the time series. Indeed, outbreaks of EHV-1 that were present in TS0 were mainly reported during winter, which is consistent with reports of seasonal patterns of disease outbreaks from a recent consensus statement [Reference Lunn38]. Removing these outbreaks from the TS0 data decreased the impact of season on the baseline and improved the smoothing performance of the two forecasting methods tested using TS1, TS2 and TS3. GLMs always provided better detection performances than HW except when using TS1. The higher specificity of TS1 compared to other time series might be explained by the positive trend and the resulting values of the variable ‘histmean’, which were smaller in TS1 than in TS0, TS2 and TS3. These results highlight the fact that pre-processing methods have an impact on the choice of best detection algorithms. However, TS0 obtained similar detection performances to time series in which outbreaks had been removed. It might be explained by the fact that data from 2011 to 2013 used to simulate our 1500 baselines were raw data that contained positive equine herpesvirus cases. Such an approach may decrease the detection performances of our outbreak-free time series (i.e. TS1, TS2, TS3) compared to raw data (TS0), but it also provides an estimation of system performances under more realistic circumstances. Our detection performances must be thus interpreted as performances for WNV outbreak detection and not as performances for the detection of WNV and equine herpesvirus. Finally, in our study, the removal of aberrations from raw data was useful to improve our models but not to improve WNV outbreaks detection performances. To our knowledge, this is the first time that the impact of past aberration removal is considered in syndromic surveillance and further work should be conducted to explore the impact and usefulness of such work.
This is the first time that an assessment of system performance has been implemented for WNV surveillance using simulated data and real data. In previous studies, assessment of timeliness, sensitivity and specificity of surveillance have occasionally been evaluated but only based on a limited number of real WNV outbreaks [Reference Calzolari8, Reference Chaintoutis39–Reference Faverjon44] which did not allow any conclusions to be drawn regarding overall system performance as also highlighted by Saegerman et al. [Reference Saegerman19]. We believe that our study helps to fill this gap and we hope that it will promote the development of such clinical surveillance system which might be one of the most cost-efficient systems for WNV early detection [Reference Saegerman19, Reference Chevalier, Lecollinet and Durand20]. Moreover, it is still difficult to identify specific clinical signs for WNV suspicion [Reference Leblond and Lecollinet45]. Promoting a surveillance system able to deal with unspecific signs, like nervous signs, would be thus especially relevant for WNV early detection.
In our study, results obtained with authentic data were similar to those obtained with simulated data: all algorithms tested were able to detect WNV outbreak in autumn 2015, the GLM always outperformed the HW method except for TS1, and specificities using simulated and real data were close. However, the number of false alarms per year estimated with simulated data was higher than the number of false alarms observed with raw data from 2011 to 2013. This is consistent with the fact that the simulated time series had more frequently high counts than raw data. In further work, it would be thus interesting to test and assess different methods for simulating time series. In addition, median timeliness and number of false alarms per year obtained with simulated data were much better than performances obtained with the authentic data. It might be explained by the specific course of WNV French outbreak in 2015 where the number of suspicions reported during the first weeks was low compared to previous WNV French outbreaks especially in 2000 [Reference Murgue13, Reference Leblond, Hendrikx and Sabatier18]. In addition, the number of cases declared to RESPE was low compared to the total number of real suspicions as, during this outbreak, the majority of cases were declared to other institutions [Reference Bahuon15]. In order to use RESPE data as an early detection system for WNV, it would be necessary to reinforce awareness of veterinary practitioners and horse-owners, and to simplify the declaration process to encourage declarations of suspect cases. Representativeness of reported syndromes is a key point in syndromic surveillance and we believe that our study will contribute to strengthen awareness of French stakeholders increasing RESPE representativeness for WNV detection.
To conclude, data on nervous signs in horses collected by RESPE can be used for the early detection of WNV outbreaks in France. As such surveillance system is based on unspecific clinical signs, it can be an efficient way to complement the official French notification system where each WNV suspect must be reported. The RESPE network is not yet fully a part of the French official diseases surveillance system even if strong links already exist between RESPE and the French ministry. Building a more integrated system including human practitioners would be valuable for both animal and human protection especially because equines may be infected by WNV before humans. In this study, we did not determine which alarm threshold was the most efficient. Such a decision would be made in real life by decision makers (e.g. RESPE, official veterinary services, public health or all together) and could be determined using cost-efficiency analysis. However, the optimal alarm threshold would also depend on the objectives of the surveillance which could be to increase public awareness for protecting human and animal health (e.g. advice to protect against mosquitoes bites, promote reporting of suspect cases), or to implement early protective measures like vector control or horse vaccination. Vaccination of horses is interesting regarding protection of animals and it was proposed during the French outbreak in 2015 in the Camargue area. However, few horses were vaccinated at that time because of the vaccine cost to the owner. The drawback of vaccination is to compromise the role of equines as a sentinel for WNV. Similarly, horses living in an area considered as endemic for WNV, like the Camargue area, may also have a long-lasting immunity and might not be efficient as sentinels for the virus. Evaluating the seroprevalence of WNV in equine populations would therefore be useful in determining which populations should receive extended vaccination coverage, or which populations living in an area considered as endemic, could still be used for early warning using a syndromic surveillance system based on clinical signs.
SUPPLEMENTARY MATERIAL
For supplementary material accompanying this paper visit https://doi.org/10.1017/S0950268816002946.
ACKNOWLEDGEMENTS
Data on nervous signs in horses were provided by RESPE (Réseau d'Epidémio-Surveillance en Pathologie Equine). The authors thank all persons involved in this network (local veterinary laboratories, and veterinarians) for collecting these data, and RESPE for providing this.
This research was performed within the VICE project funded under the FP7 EMIDA ERA-NET initiative. National funding within ERA-Net was provided by the Dutch Ministry of Economic Affairs (Project no. BO-20-009-009). This work was also partially financially supported by IFCE (Institut Français du Cheval et de l'Equitation) and Vetagro Sup (National Veterinary School of Lyon).
DECLARATION OF INTEREST
None













