Methicillin-resistant Staphylococcus aureus (MRSA) is a major cause of hospital-associated (HA-MRSA), community-associated (CA-MRSA) and livestock-associated (LA-MRSA) infections [Reference Otter and French1]. In 2011, a novel divergent mecA gene homologue (mecALGA251), designated mecC, was discovered. This gene has less than 70% homology with the mecA gene and is associated with a novel staphylococcal cassette chromosome (SCCmec) type XI [Reference Peterson2].
MRSA isolates harbouring the mecC gene have been reported in several European countries mainly from humans who had contact with livestock, and/or wild and domestic animals [Reference Peterson2, Reference Paterson, Harrison and Holmes3]. Although the isolates with the mecC gene are associated with livestock, they differ from LA-MRSA [clonal complex (CC) 398] isolates related to pigs, which are highly resistant to tetracyclines used in pig production [Reference Peterson2]. The range of infections caused by mecC-carrying MRSA is the same as seen in other S. aureus, including life-threatening diseases such as bacteraemia [Reference Otter and French1, Reference Paterson, Harrison and Holmes3].
MRSA is well-controlled in Slovenian hospitals. Some documented outbreaks include four cases of skin and soft tissue infections due to a CA-MRSA strain obtained from one hospital in 2003 and 2004 (spa type t044, sequence type (ST)80) [Reference Dermota4] and in 2005, Panton–Valentine leukocidin (PVL)-positive CA-MRSA strains were identified in football players (spa type t002, ST5 and spa type t454, ST152) [Reference Műller-Premru5]. To date, mecC-positive MRSA isolates in animals, humans, or persons having direct contact with animals have not been reported or documented in Slovenia.
Case description and epidemiological investigation
The first mecC-positive MRSA was isolated from an 86-year-old female inpatient in a regional hospital in northeastern Slovenia, who was hospitalized after a stroke in April 2013. Although the patient had no risk factors for nosocomial acquisition in the previous year, namely hospitalization history or surgery, use of an indwelling catheter or other medical devices, and did not show any signs of infection, she was screened for MRSA upon admission. Nose and skin swabs taken within 48 h after admission were MRSA positive. A screening test for methicillin resistance (30-μg cefoxitin disk on Mueller–Hinton II agar; BD, USA) categorized the strain as resistant, but a slide agglutination assay for PBP2a/PBP2’ (Oxoid) and PCR for the mecA gene [Reference Dermota4] were negative. Due to these discrepancies, PCR for mecC gene was performed [Reference Peterson2] which proved positive. The index patient lived on a farm and had contact with pigs and companion animals (cats, dog), but not with cattle and sheep. Two months after the MRSA isolation in the index patient an epidemiological investigation including sampling of the relatives, animals and farm environmental samples was performed. Throat and nose swab were taken again from the patient and from all members of the family (husband, daughter, son-in-law). Nasal swabs were taken from seven clinically healthy piglets and dust samples from their environment. All the samples were analysed according to the protocol used in a baseline study on MRSA in holdings of breeding pigs to assess the prevalence and diversity of MRSA in pig primary production [6].
Only the index patient was again positive for mecC MRSA, while all other samples from humans, animals and the environment were negative. Therefore the source and origin of the index patient's mecC-positive MRSA isolate remained unclear.
Screening for mecC-positive strains in a CA-MRSA collection
Among MRSA carrying the mecC gene, antimicrobial susceptibility patterns are most similar to CA-MRSA, in being classically susceptible to the majority of non-β-lactam antibiotics [Reference Grmek Košnik7]. Clinical data also indicate that mecC MRSA are primarily community-associated. As we do not routinely screen for mec genes in phenotypically confirmed MRSA in Slovenia, we therefore sought to identify additional mecC strains through a retrospective screen survey of mec genes in the Slovenian national collection of presumptive CA-MRSA isolates. This collection is maintained at the National Laboratory for Health, Environment and Food (NLZOH) and currently contains 395 isolates recovered since 2006. Inclusion criteria for the presumptive CA-MRSA collection were based on phenotypic definition of being resistant to cefoxitin and oxacillin, and susceptible to at least two of the following four antibiotics: ciprofloxacin, erythromycin, clindamycin or gentamicin. All isolates in the collection were further genotypically characterized by SCCmec type, presence of PVL, and spa type. The majority of isolates met the latest relevant molecular definition of CA-MRSA [Reference Otter and French1]. No replicate MRSA isolates from the same patient were included.
The mecA gene was detected in 385 (97·5%) isolates while 10 (2·5%) were mecA-negative. Six of these 10 were positive for the mecC gene. The remainder remain under investigation of the genetic basis of β-lactam resistance. Two of the mecC-positive MRSA strains were isolated from healthy carriers during routine surveillance for MRSA and four were recovered from clinical specimens from wound, skin and soft tissue infections. The oldest mecC-positive MRSA isolate was from 2007.
Susceptibility to antibiotics was tested using a standardized agar disk diffusion method according to Clinical Laboratory Standards Institute (CLSI) guidelines [8]. The mecC strain from the index patient and all six strains from the collection were resistant to penicillin and cefoxitin, but susceptible to vancomycin, gentamicin, tobramycin, kanamycin, erythromycin, clindamycin, tetracycline, ciprofloxacin, trimethoprim–sulfamethoxazole, chloramphenicol, rifampin, mupirocin and fusidic acid. The minimum inhibitory concentration (MIC) for oxacillin was performed using the E-test (bioMérieux, France). The strain from the index patient displayed an oxacillin MIC of 24 mg/l, and one strain from the collection was 2 mg/l; the remainder were between 4 and 16 mg/l.
The majority of mecC-positive MRSA strains originated from rural areas of northeastern and southern Slovenia, and only one strain originated from the western region of Slovenia. Epidemiological information about animal contact in these patients was lacking. The clustering of cases in rural areas and not in urban regions indicates that contact with livestock could be a risk factor [Reference Otter and French1, Reference Paterson, Harrison and Holmes3].
Molecular characterization of mecC MRSA strains
All mecC MRSA strains were investigated by DNA microarray using StaphyType kit 2.0 (Alere Technologies GmbH, Germany) to detect genes encoding species markers, antimicrobial resistance genes, virulence genes and typing markers (SCCmec, capsule, agr) at the French National Reference Centre for Staphylococci in Lyon [Reference Monecke9], and were spa-typed [Reference Harmsen10]. The characteristics of all seven mecC strains are shown in Table 1. The genes encoding PVL and lukM, toxic shock syndrome toxin, exfoliative toxins, ACME, enterotoxins and genes sak, chp, scn were absent. The lack of the latter genes which are involved in human immune evasion, in all seven strains could indicate their possible adaptation to animals rather than humans [Reference Peterson2, Reference Paterson, Harrison and Holmes3]. None of the mecC strains carried genes for resistance to other antibiotics.
Table 1. Characterization of seven mecC-positive MRSA isolated from humans in Slovenia, 2006–2013
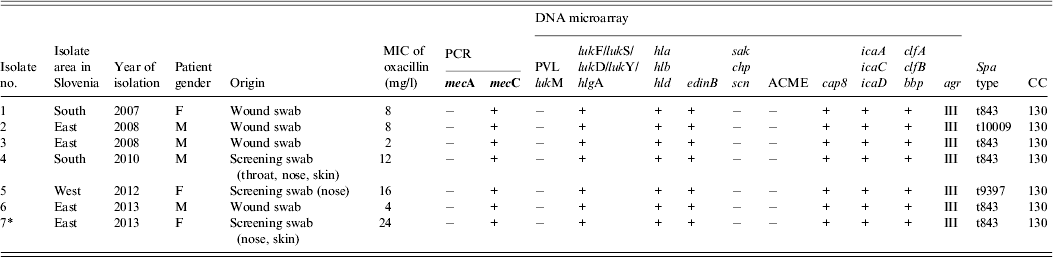
* Index patient.
F, female; M, male; MIC, minimum inhibitory concentration; PCR, polymerase chain reaction; PVL, Panton–Valentine leukocidin; lukM, leukocidin M; lukF, leukocidin F; lukS, leukocidin S; lukD, leukocidin D; lukY, leukocidin Y; hlg, haemolysin gamma; hla, haemolysin alpha; hlb, haemolysin beta; hld, haemolysin delta; edinB, epidermal cell differentiation inhibitor B; sak, staphylokinase; chp, chemotaxis inhibitory protein; scn, staphylococcal complement inhibitor; ACME, arginine deaminase; cap8, capsule type 8; icaA, intracellular adhesion protein A; icaC, intracellular adhesion protein C; icaD, biofilm PIA synthesis protein D; clfA, clumping factor A; clfB, clumping factor B; bbp, bone sialoprotein-binding protein; agr, accessory gene regulator; CC clonal complex.
A range of sequence types and clonal complexes have been identified in mecC strains from humans and a diverse range of animal species throughout Europe (CC49, CC130, CC425, CC599, CC1943) [Reference Peterson2, Reference Paterson, Harrison and Holmes3]. All our strains belonged to CC130 and to three different spa types, t843 (n = 5), t9397 (n = 1) and t10009 (n = 1). Similar results have been observed in other studies [Reference Otter and French1, Reference Paterson, Harrison and Holmes3].
In conclusion, MRSA strains positive for mecC have been present in Slovenia since 2007, but were only recognized in routine laboratory testing for MRSA in 2013. Because the oxacillin MIC can be below the cut-off point for resistance, and in the susceptible range of the CLSI breakpoint (⩽2 mg/l), these MRSA strains may be overlooked. All microbiology laboratories should be aware of the possibility of mecC S. aureus and isolates resistant or intermediately resistant to oxacillin or cefoxitin and mecA-negative should be tested with an appropriate PCR for the mecC gene. Failure to detect these MRSA strains could have serious consequences on public health. Finally, ongoing surveillance for mecC MRSA in humans, animals, food and persons in close contact with animals is required to detect changes in MRSA epidemiology in Slovenia.
ACKNOWLEDGEMENTS
The authors thank Martina Kavčič, Slavica Lorenčič-Robnik, Tatjana Harlander, Nataša Švent-Kučina, Jerneja Fišer, Ljudmila Sarjanovič and Tjaša Žohar-Čretnik for providing the presumptive CA-MRSA strains to the national collection.
DECLARATION OF INTEREST
None.




