Introduction
Bordetella bronchiseptica can infect humans and a number of animal species [Reference Guiso and Hegerle1]. The infection of B. bronchiseptica in immunocompetent humans is rare. However, the pathogen was reported to associate with respiratory diseases in immunosuppressed humans [Reference Yacoub2–Reference Lorenzo-Pajuelo4]. In animals, the pathogen is also associated with respiratory diseases, such as canine infectious respiratory disease syndrome in dogs, tracheobronchitis in cats, atrophic rhinitis in pigs, pneumonia in horses and snuffles in rabbits [Reference Chambers5–Reference Deeb9]. It has been showed that humans could be infected with B. bronchiseptica by direct contact with infected animals (swine, dog, rabbit and cat), causing diseases including whooping cough, bronchitis, bronchopneumonia, pneumonia and peritonitis [Reference Woolfrey and Moody10–Reference Register12]. Therefore, more efforts should be endeavoured to understand the epidemiology and characteristics of B. bronchiseptica in animals, which will help in preventing the transmission of B. bronchiseptica from animals to humans.
Fujian Province, in the southeast of China, is one of the important rabbit-farming areas in China [Reference Wang13]. The number of live rabbits farmed was about 9.4 million by the end of 2018 (Fujian Statistical Yearbook, 2019). In Fujian Province, all the rabbits are used for food, and the rabbit meat production reached 14.1 thousand tonnes in 2018 (Fujian Statistical Yearbook, 2019). Traditionally, the hair and the abdominal organs (except for heart and liver) of the rabbit are removed and the slaughter rate is around 75%.
B. bronchiseptica spreads rapidly in rabbits and the infection is often persistent [Reference Liu14]. To the best of our knowledge, B. bronchiseptica is a common pathogen isolated from rabbits with respiratory diseases in Fujian Province. However, information on the prevalence and characteristics of B. bronchiseptica in rabbits in Fujian Province is limited. In this study, B. bronchiseptica strains were recovered from the lung samples of dead rabbits with respiratory diseases. The isolates were characterised by multi-locus sequencing typing, screening the virulence genes and testing the antimicrobial susceptibility.
Methods
Ethics statement
Lung samples were collected from dead rabbits with respiratory diseases in accordance with the guidelines issued by the Research Ethics Committee of the Institute of Animal Husbandry and Veterinary Medicine, Fujian Academy of Agriculture Sciences (FAAS). This study was also approved by the Research Ethics Committee of the Institute of Animal Husbandry and Veterinary Medicine, and the approved number is FAAS-AHVM2017-0511.
Sample collection and B. bronchiseptica isolation
Nine rabbit farms in Fuzhou, Longyan and Nanping cities in Fujian Province were included for sampling. The two rabbit farms in Fuzhou raise Fujian Yellow rabbit (local rabbit breed), the three rabbit farms in Longyan raise Fujian White rabbit (local rabbit breed), the other two rabbit farms in Longyan raise Minxinan Black rabbit (local rabbit breed) and the two rabbit farms in Nanping raise Hyplus rabbit (foreign rabbit breed). The whole lungs were collected from dead rabbits with respiratory diseases. In all, 833 whole lungs were collected from August 2017 to December 2019.
In order to isolate B. bronchiseptica, 1 g lung tissue (lower lobe) from each whole lung was mixed with 1 ml of sterile phosphate buffer saline and homogenised to make 50% suspension. One hundred microlitres of suspension was spread on brain heart infusion (BHI) agar plate (containing 1% glycerol and 10% defibrinated sheep blood) and incubated for 48–72 h at 37 °C. The suspected smooth, round, greyish white and shiny colonies with the diameter around 1 mm were picked up and purified. Five isolates of Gram negative, oxidase and ornithine decarboxylase positives, and glucose fermentation negative were further confirmed by sequencing of the 16S rRNA genes. One isolate from each lung sample was selected as representative for further characterisation.
Multi-locus sequence typing
The isolates were analysed by multi-locus sequence typing (MLST) as described in PubMLST website (https://pubmlst.org/). Briefly, seven house-keeping genes including adk, fumC, glyA, tyrB, icd, pepA and pgm were amplified and sequenced. The seven allelic numbers were given by comparing the seven house-keeping genes of the isolate to the corresponding genes in the PubMLST database. The sequence types (STs) of the isolates were defined according to the seven allelic numbers.
Virulence gene detection
Five well-characterised virulence genes that thought to be involved in interactions with the host were screened in the isolates, including filamentous haemagglutinin (FHA) (fhaB), pertactin (prn), adenylate cyclase-haemolysin toxin (cyaA), dermonecrotic toxin (dnt) and the Bordetella type-III secretion system effector A (bteA) [Reference Cotter15–Reference Kuwae19]. The primers for amplification of the five virulence genes were designed based on the corresponding genes in the NCBI database (https://www.ncbi.nlm.nih.gov/) (Table 1). Polymerase chain reaction (PCR) mixtures comprised of 25 μl 2× EasyTaq PCR SuperMix (TransGen Biotech, Beijing, China), 0.2 μM of each forward and reverse primer and 100 ng bacterial genome DNA in a final volume of 50 μl. The cycling conditions for PCR assays were 94 °C for 5 min and 35 cycles of 94 °C for 30 s, 59 °C for 30 s and 72 °C for 30 s, followed by a final elongation step of 72 °C for 5 min. The B. bronchiseptica strain MH001 and the sterile ddH2O were included as the positive and negative controls, respectively [Reference Wang20]. The PCR products were sequenced to confirm the identities.
Table 1. Primers used for amplification of the five virulence genes in the B. bronchiseptica isolates

Antimicrobial susceptibility test
The antimicrobial susceptibility test of the isolates were conducted by using disc diffusion method on BHI agar plate (containing 1% glycerol and 10% defibrinated sheep blood) according to the Clinical and Laboratory Standards Institute (CLSI) standards. Ten antibiotics with the stated concentrations (μg/disc) were included: ampicillin (10), cefixime (5), cefotaxime (30), ceftizoxime (30), gentamycin (10), kanamycin (30), streptomycin (10), ofloxacin (5), ciprofloxacin (5) and norfloxacin (10). The results were interpreted based on the breakpoints for Enterobacteriaceae of the CLSI standards [21, Reference Loong22]. The Staphylococcus aureus ATCC 29213 was included as the quality control.
Results
B. bronchiseptica isolation and identification
Two hundred and nineteen B. bronchiseptica isolates were recovered from the 833 whole lung samples of dead rabbits with respiratory diseases. The B. bronchiseptica isolates could be recovered from the all nine rabbit farms and the all four rabbit breeds (Table 2). The infection rates of B. bronchiseptica in the nine rabbit farms and the four rabbit breeds ranged from 13.79% to 38.42% and 15.66% to 50.24%, respectively.
Table 2. Distribution, origin and STs of the 219 B. bronchiseptica isolates
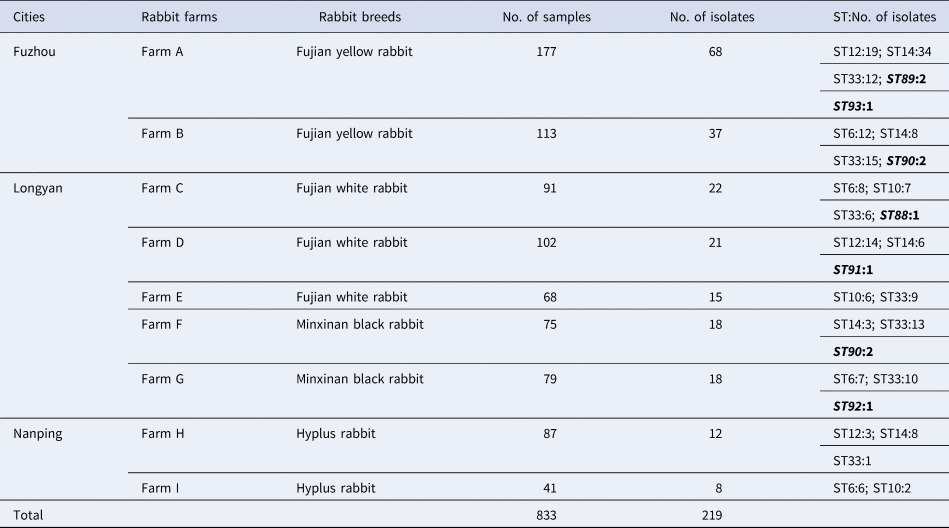
The bold-italic characters indicate the new STs.
Multi-locus sequence typing
The results of MLST analyses showed that the 219 B. bronchiseptica isolates were typed into 11 STs, with the ST33 (30.14%, 66/219), ST14 (26.94%, 59/219) and ST12 (16.44%, 36/219) were the three most predominant STs. Among the 11 STs, six new STs were detected including ST88, ST89, ST90, ST91, ST92 and ST93 (Table 2), and the MLST profiles of the six new STs had been submitted to the PubMLST database (https://pubmlst.org/bigsdb?db=pubmlst_bordetella_seqdef&page=profiles&scheme_id=3). The seven house-keeping genes of each of the isolate of the 11 STs were concatenated for phylogenetic analysis. A maximum likelihood tree was constructed (no. of bootstrap replications was 1000) by using the MEGA 5.0 software. The 11 STs were mainly grouped into two clusters, cluster I and cluster II. Cluster I contained nine STs including ST6, ST10, ST12, ST14, ST33, ST88, ST89, ST90 and ST91, whereas cluster II only contained two STs including ST92 and ST93 (Fig. 1).
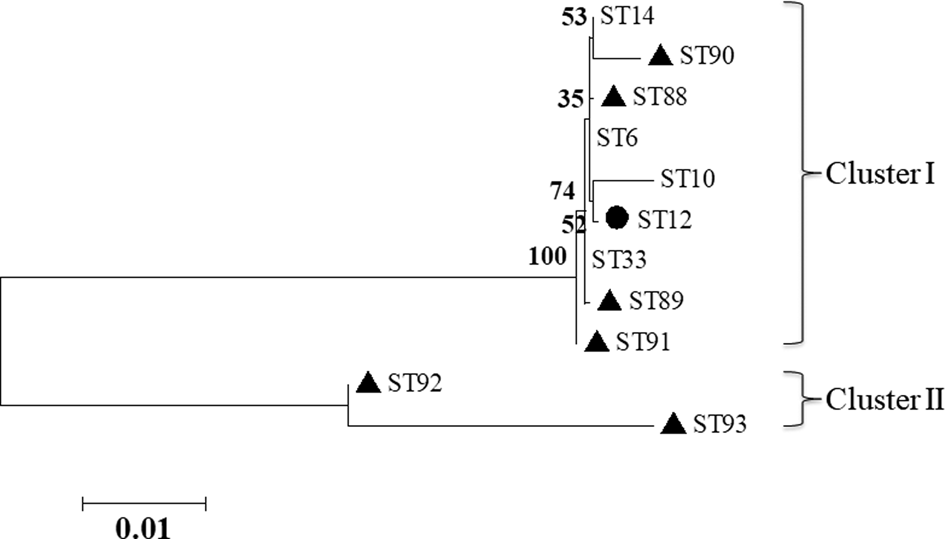
Fig. 1. Phylogenetic tree of the 11 STs of B. bronchiseptica based on the concatenated seven house-keeping genes. The black filled triangles indicate the new STs. The black filled circle indicates the B. bronchiseptica strain of ST12 that can be isolated from both rabbits and humans.
Virulence gene detection
Five virulence genes were screened in the 219 B. bronchiseptica isolates. The results showed that all the five virulence genes of fhaB, prn, cyaA, dnt and bteA were positive in the all 219 isolates.
Antimicrobial susceptibility test
The results of antimicrobial susceptibility test showed that most of the B. bronchiseptica isolates in this study were susceptible or intermediate susceptible to streptomycin, ciprofloxacin, gentamycin, ofloxacin, kanamycin and norfloxacin, and the sensitive rates to the six kinds of drugs were 100%, 99.09%, 98.63%, 96.80%, 96.34% and 21%, respectively (Table 3). The isolates resistant to cefixime, ceftizoxime, cefatriaxone and ampicillin were observed, and the resistance rates to these four kinds of drugs were 33.33%, 31.05%, 11.87% and 3.20%, respectively (Table 3). No isolate of multi-drug resistant was detected.
Table 3. Antimicrobial susceptibility test of the 219 B. bronchiseptica isolates
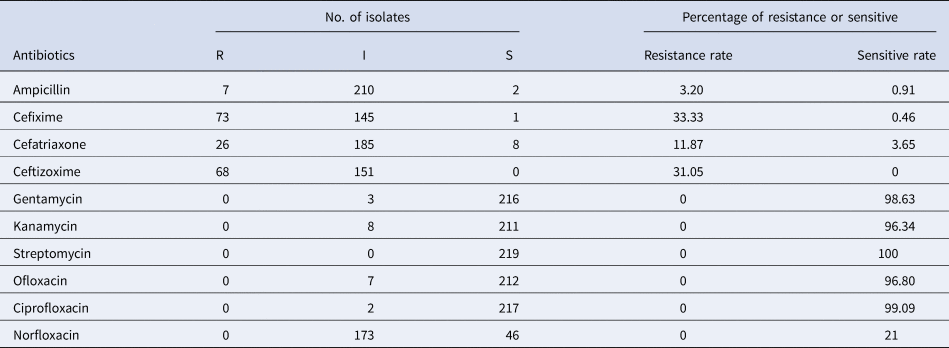
‘R’ represents resistance; ‘I’ represents intermediate; ‘S’ represents susceptible.
Discussion
The present study described the epidemiology and characteristics of B. bronchiseptica in rabbits in Fujian Province. The results showed that B. bronchiseptica was prevalent in the nine rabbit farms and the four rabbit breeds, and the infection rate of B. bronchiseptica in rabbits in Fujian Province was as high as 26.29% (219/833). The results suggested that B. bronchiseptica is an important pathogen causing respiratory diseases in rabbits in Fujian Province.
The 219 B. bronchiseptica isolates in this study were typed into five known STs (ST6, ST10, ST12, ST14 and ST33) and six new STs (ST88, ST89, ST90, ST91, ST92 and ST93). In the PubMLST database (https://pubmlst.org/bordetella/), B. bronchiseptica strains of ST6 are isolated from rabbits in Switzerland, strains of ST10 are isolated from rabbits in USA, strains of ST12 are isolated from rabbits in USA and Denmark, and strains of ST14 are isolated from rabbits in USA and UK. However, strains belonging to ST33 that the most prevalent ST (30.14%, 66/219) in rabbits in Fujian Province are isolated from seal, dog, leopard and horse, but not from rabbits. Notably, strains of ST12 could also be isolated from humans, suggesting the potential zoonotic transmission between rabbits and humans [Reference Gueirard11]. Moreover, the five known STs (ST6, ST10, ST12, ST14 and ST33) are grouped into the clonal complex of ST6 complex (the ST6 is the putative founder) in the PubMLST database. By comparing the MLST profiles of the six new STs (ST88, ST89, ST90, ST91, ST92 and ST93) with that of ST6, ST88 showed the most genetic closely to ST6 (ST88 is the single locus variant of ST6) [Reference Francisco23].
Expression of the virulence factors facilitates the invasion of B. bronchiseptica in hosts [Reference Fingermann and Hozbor24]. Surprisingly, all the 219 B. bronchiseptica isolates in this study carried the five screened virulence genes of fhaB, prn, cyaA, dnt and bteA. The fhaB and prn genes encode the FHA and pertactin (PRN), respectively. The FHA and PRN are important adhesins expressed in the genetic closely related species of B. pertussis, B. parapertussis and B. bronchiseptica, which mediates the bacterial adhesion of host cells [Reference Parkhill25, Reference Edwards, Groathouse and Boitano26]. It should be aware that the PRN-deficient B. pertussis had emerged, and the losses of the prn gene expression in the strains might be driven by using of acellular vaccine containing PRN [Reference Carriquiriborde27]. The cyaA gene encodes the multifunctional adenylate cyclase-haemolysin (AC-Hly) toxin, which possess adenylate cyclase activity, pore-forming activity and cell invasive activity [Reference Guiso17]. It was showed that the AC-Hly toxin expressed in the B. bronchiseptica strains isolated from human and rabbit was responsible for the lethality of the intranasally infected mice [Reference Gueirard11]. The dnt gene encodes the dermonecrotic toxin (DNT), and the toxin is a well-recognised causative factor inducing atrophic rhinitis and bronchopneumonia in pigs [Reference Brockmeier18]. Interestingly, the ability to express DNT is varied among strains. It was showed that the larger amount of DNT was produced in pig isolates than in a rabbit isolate RB50 [Reference Okada28]. The bteA, also known as bopC, encodes the Bordetella type III secretion system effector A (BteA), which is an important effector secreted from the type III secretion system [Reference Kuwae19, Reference Kuwae29]. It was showed that BteA could induce the necrotic cell death and inhibit the macrophage phagocytosis [Reference Kuwae19, Reference Kuwae29]. Taking together, the 219 B. bronchiseptica isolates carrying the five virulence genes in this study were the potential pathogen causing severe respiratory infection in rabbits.
Antibiotics play an important role in prevention and treatment of infections caused by B. bronchiseptica [Reference Lappin30–Reference Kadlec, Kehrenberg and Schwarz32]. However, the widespread use of antibiotics for preventing and treating the bacterial infections leads to the emergence of antibiotic-resistant strains [Reference Kadlec, Kehrenberg and Schwarz32–Reference Kadlec, Kehrenberg and Schwarz34]. It was demonstrated that B. bronchiseptica strains isolated from swine in Germany were resistant to ampicillin, and B. bronchiseptica strains isolated from companion animals in Germany and other European countries showed decreased susceptibility to β-lactam antibiotics and cephalosporins [Reference Prüller35]. In consistence with this study, B. bronchiseptica isolates showing resistant to ampicillin and cephalosporins were also observed in this study. It was showed that the production of beta-lactamases and the reduced membrane permeability to cephalosporins might result in the resistance to ampicillin and cephalosporins, respectively [Reference Kadlec36]. Therefore, it should be aware that the widespread use of antibiotics to control the infection caused by B. bronchiseptica in rabbits is unsustainable.
The characteristics of the B. bronchiseptica strains isolated from dead rabbits with respiratory disease in Fujian Province were described in this study. The results showed that B. bronchiseptica was an important pathogen causing respiratory diseases in rabbits in Fujian Province, and that B. bronchiseptica stain of ST12 that can infect humans was also isolated from rabbits in Fujian Province. Unexpectedly, B. bronchiseptica isolates belonging to six new STs (ST88, ST89, ST90, ST91, ST92 and ST93) were detected, and B. bronchiseptica isolates resistance to cefixime, ceftizoxime, cefatriaxone and ampicillin were also detected. The results might play important roles in tracking the epidemic strains in rabbits, controlling the B. bronchiseptica infections in rabbits and preventing the potential rabbit–human transmission events.
Financial support
This work was supported by the Fujian Public Welfare Project (2019R1026-9) and National Rabbit Industry Technology System of People's Republic of China (CARS-43-G-5).
Conflict of interest
None.
Data availability statement
Requests for access to the data that support this study should be made to the corresponding author, XX.







