Introduction
The epidemiology of methicillin-resistant Staphylococcus aureus (MRSA) continues to evolve. With the exception of a few countries, the overall prevalence of MRSA has increased around the world owing to inevitable evolution of the microorganism propelled by ubiquitous antimicrobial pressure [Reference Stefani and Varaldo1–3]. Various epidemiologically important MRSA strain types have been described, defined by the type staphylococcal cassette chromosome (SCCmec) genetic elements and unique pulsed-field gel electrophoresis (PFGE) patterns [Reference Hiramatsu4–Reference Tenover6]. In Canada, ten epidemic MRSA strain lineages have been identified based on PFGE analysis, designated CMRSA1 to CMRSA10 [Reference Simor7, Reference Christianson8]. CMRSA10 and 7 are considered community-associated (CA-MRSA) strains while the others, mainly CMRSA2, are primarily healthcare-associated strains (HA-MRSA).
Although MRSA remains a significant nosocomial pathogen, more concerning has been the rapid emergence of CA-MRSA causing infections primarily among those without significant healthcare contacts [9–Reference Boucher and Corey11]. Specifically, CMRSA10 (USA300) has been the predominant strain of CA-MRSA in North America causing the majority of skin and soft-tissue infections in the community and an increasing proportion of bloodstream infections in the healthcare setting [Reference Moran12–Reference Klevens14]. A recent report from the Canadian Nosocomial Infection Surveillance Program showed that the proportion of CMRSA10 among nosocomial infections was on the rise, highlighting its migration into the healthcare setting [Reference Simor15].
In 2004, an outbreak of CMRSA10 occurred in Alberta in a marginalized urban population with histories of intravenous drug use, homelessness, and incarceration [Reference Gilbert16]. This outbreak and the increasing concern of spread of MRSA into the general population prompted enhanced province-wide MRSA surveillance in Alberta. We report the results of a prospective population-based surveillance of MRSA infection describing the evolving molecular epidemiology of MRSA in Alberta over a consecutive 3-year period.
Methods
Surveillance population
Alberta is a province in western Canada with a population of approximately 3·6 million. Until 2009, healthcare delivery was regionalized across nine health authorities with regional laboratories providing diagnostic microbiology services to each health region. These regions represent the Calgary and Edmonton areas, Northern, Central and Southern Alberta (Fig. 1). The Alberta Provincial Laboratory for Public Health (ProvLab) provides province-wide reference and molecular-typing services for targeted notifiable pathogens. From 1 June 2005, MRSA was designated a ‘pathogen under surveillance’ across the province and all regional laboratories were notified by the Chief Medical Officer of Health to submit all first MRSA clinical isolates to the ProvLab for molecular typing.
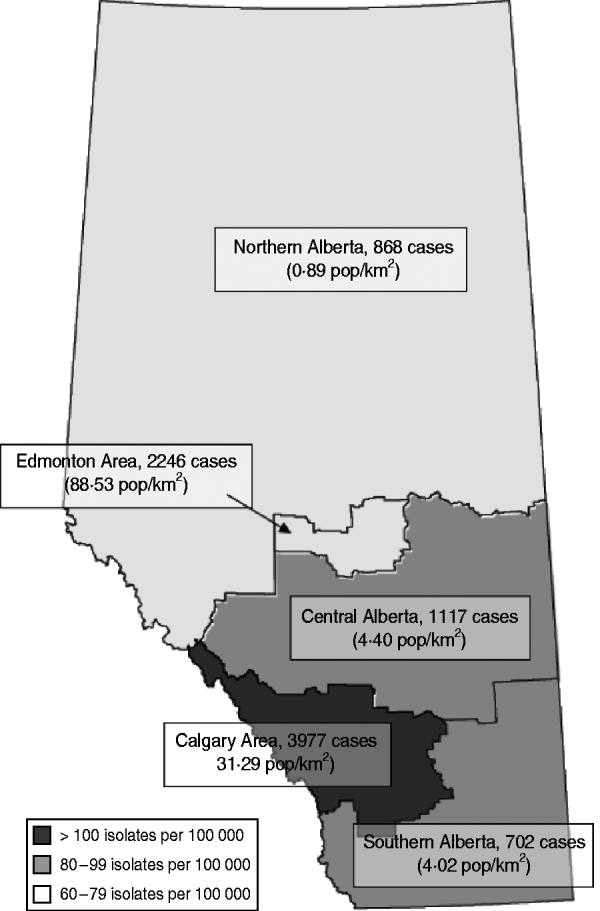
Fig. 1. Total number of isolates and estimated overall annual incidence of methicillin-resistant Staphylococcus aureus in Alberta, 2005–2008. Based on 3-year total populations of Northern Alberta, Edmonton Area, Central Alberta, Calgary Area, and Southern Alberta. Population density displayed as average of years 2005–2008.
Isolate collection and classification
Regional laboratories performed testing on all cultures collected as deemed necessary by the physicians and consisted of clinical specimens obtained for diagnostic purposes as well as screening specimens. A ‘first clinical isolate’ was defined as an isolate detected from a patient without previous MRSA isolates in the preceding 12 months. Screening isolates were excluded from the study. A case of MRSA was defined for each clinical isolate. Regional laboratories, as per their routine procedures, performed detection and confirmation of MRSA. Each isolate was then submitted to the ProvLab along with a MRSA isolate submission form containing the following data elements: (1) specimen source information including date of specimen collection and submitting physician; (2) patient demographic information and clinical diagnosis; and (3) specimen clinical information including specimen collection location (in-patient, outpatient, long-term care facility, correctional facility, military, drop-in centre, or other), MRSA isolate (screening, clinical, referred by Medical Examiner's Office, or unknown), anatomical specimen site, and previous MRSA positivity in the past 12 months.
The ProvLab entered all data in the provincial laboratory information system. For the purpose of this study, all first MRSA clinical isolates collected from 1 July 2005 to 30 June 2008 were included. The database was re-examined to confirm a single clinical isolate per patient using at least two matching unique identifiers: personal health number, name, and/or date of birth. If two or more isolates were collected from the same patient, only the first clinical specimen separated by at least 12 months was accepted for analysis. The source of clinical isolates was classified into the following: blood; sterile (joints/synovial fluid, pleural fluid, cerebrospinal fluid, any internal organ tissue or bone biopsy specimen); respiratory tract (sputum, endotracheal aspirate, or bronchial specimen); urine; skin and soft tissue (surgical wounds, other anatomical areas without any descriptor); devices (including Foley catheters, devices, central venous catheter tip); or other (drainage from unspecified source or mucous membrane). Isolates obtained from blood and sterile sites were considered invasive.
Molecular typing
At the ProvLab, PFGE on MRSA isolates was performed according to standardized protocol [Reference Mulvey5] and the fingerprints generated were entered into the Alberta database. BioNumerics software version 5.0 (Applied Maths, USA) was used for the analysis. All fingerprint profiles were compared to the existing library containing previous Alberta PFGE profiles and the ten known prototype strain profiles representing the respective Canadian epidemic strain types (CMRSA1-10) [Reference Christianson8]. If the fingerprint was indistinguishable from a Canadian prototype strain, it was given a CMRSA1-10 designation. Any fingerprint showing fewer than seven bands' difference to the prototype strain was considered to be related and assigned to the respective CMRSA strain type according to the criteria as described by Tenover et al. [Reference Tenover17]. When a PFGE pattern differed by ⩾7 bands from the respective CMRSA strain type, it was designated as ‘not-assigned’ and having no relatedness with the existing Canadian epidemic clones.
Antimicrobial susceptibility testing
Antimicrobial susceptibility testing was performed on all blood isolates including those belonging to CMRSA2, 7, and 10 clones. These isolates were sent to the National Microbiology Laboratory (Winnipeg, Canada) to determine minimum inhibitory concentrations (MIC, μg/ml) against 12 antimicrobial agents (clindamycin, ciprofloxacin, erythromycin, fusidic acid, gentamicin, linezolid, mupirocin, quinupristin–dalfopristin, rifampin, tetracycline, trimethoprim–sulfamethoxazole, vancomycin). Antimicrobial susceptibility testing using microbroth dilution was performed according to Clinical Laboratory Standard Institute (CLSI) guidelines [18], with the exception of interpretation of mupirocin and fusidic acid resistance. Susceptibility to mupirocin was defined using the following MIC breakpoints: <4 μg/ml, susceptible; ⩾4 to <256 μg/ml, low-level resistance; and ⩾256 μg/ml, high-level resistance [Reference Walker19]. Fusidic acid resistance was defined as a MIC ⩾2 μg/ml [Reference Skov, Frimodt-Møller and Espersen20]. Inducible resistance to clindamycin in macrolide-resistant isolates of MRSA was determined by a standardized disk approximation test [18].
Data analysis
Incidence rates of MRSA infection were calculated by dividing the total number of MRSA clinical isolates by the mid-year population denominators obtained from Alberta Health Care Insurance Plan Stakeholder Registry and expressed as cases/100 000 population with 95% confidence intervals (CI). For 2005 and 2008, the annual incidence was estimated by doubling the number of cases to reflect 6-month rates. Trends in rates were determined using Poisson regression. Proportions among categorical data were compared using Fisher's exact test. Multiple group comparisons were made using Tukey-style multiple comparisons of proportions [Reference Zar21]. A P value of <0·05 was considered statistically significant.
Results
Incidence and demographical characteristics
Between 1 July 2005 and 30 June 2008, a total of 10 184 clinical isolates was received. After excluding 1274 isolates (duplicate results and other errors), 8910 clinical MRSA isolates, each representing a case of infection, were analysed.
There was an increase in the overall incidence of MRSA infections over the study period (Fig. 2). The rates of CMRSA10 and 7 increased significantly (P<0·05 and P<0·05, respectively), while the rate of CMRSA2 did not change over time (P=0·11). Overall, 5191 (58·3%) infections occurred in males (incidence rate ratio 1·44, 95% CI 1·39–1·49, P<0·05). The highest total number of MRSA infections occurred in the 35–44 years age group with 1528 cases, while the ⩾85 years age group had the highest overall annual incidence with 583 cases/100 000 population (Table 1). The median age was younger among those with CMRSA10 and 7 compared to those with CMRSA2 and other MRSA strain types as shown in Figure 3. Age-specific incidence rates by MRSA strain types are presented in Figure 4. The Calgary Area had the highest number of MRSA infections throughout the study period with 3977 cases and the highest overall annual incidence with 106 cases/100 000 population (Fig. 1). The two major urban centres, Calgary and Edmonton areas, accounted for 44·6% and 25·2% of total MRSA infections in Alberta, respectively.
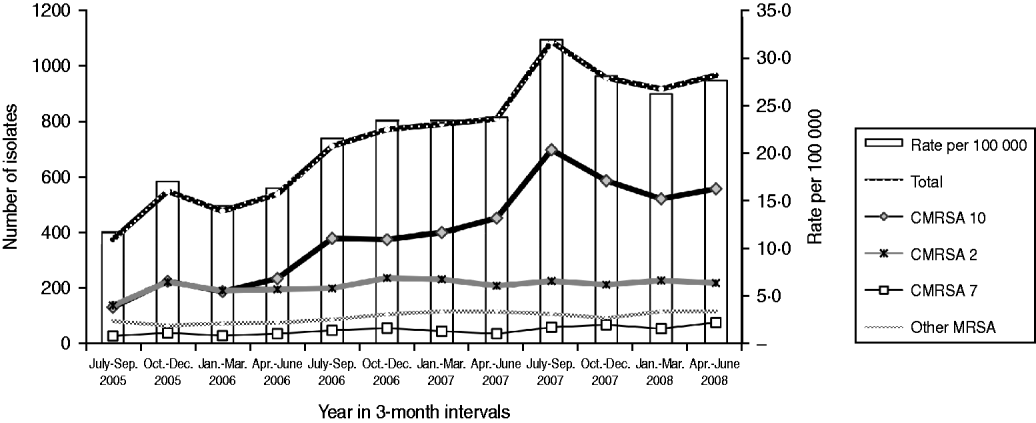
Fig. 2. Rate of methicillin-resistant Staphylococcus aureus infection in Alberta, 2005–2008.
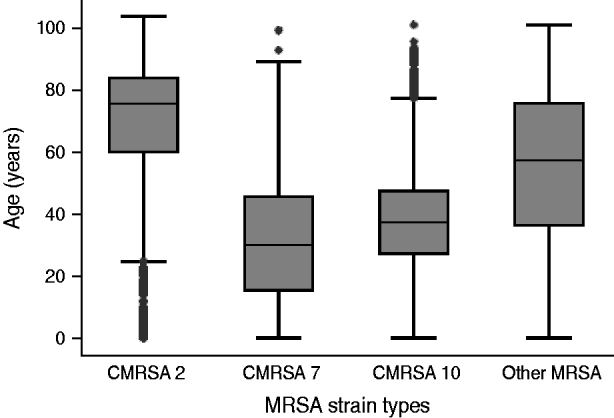
Fig. 3. Distribution of age in patients with different methicillin-resistant Staphylococcus aureus (MRSA) strain types.
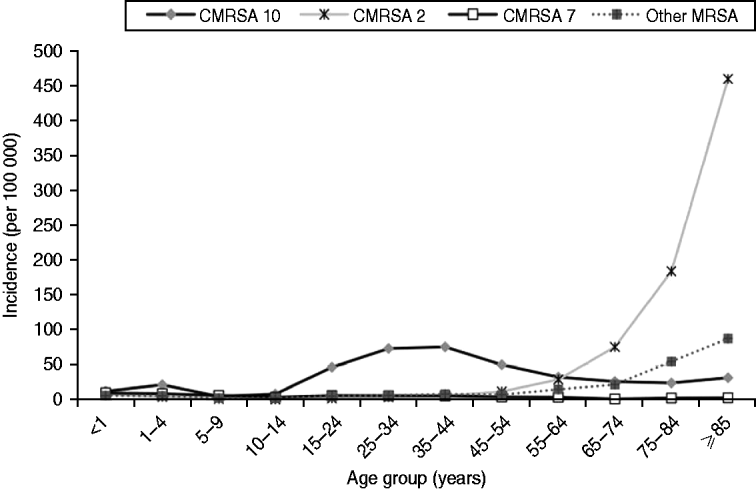
Fig. 4. Age-specific incidence (no. of isolates/100 000 per year) of methicillin-resistant Staphylococcus aureus (MRSA) by strain types in Alberta, Canada, 2005–2008.
Table 1. Estimated incidence (no. of cases per 100 000 population per year) of methicillin-resistant Staphylococcus aureus (MRSA) infections in Alberta, Canada, 2005–2008
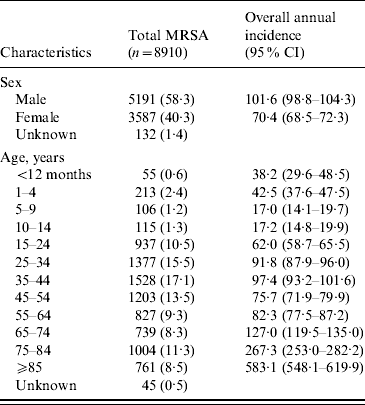
CI, Confidence interval.
Data represented as no. (%) of patients unless otherwise indicated.
Molecular typing and clinical characteristics
The most common strain type was CMRSA10 with 4723 (53·0%) cases. CMRSA2 accounted for 2488 (27·9%) cases while CMRSA7 constituted 562 (6·3%) cases. Other MRSA included other known CMRSA strain types (483, 5·4%) and MRSA isolates ‘not assigned’ by PFGE to any known Canadian epidemic prototype strain (654, 7·3%).
The number of different fingerprint patterns including the respective prototype patterns generated for MRSA isolates by PFGE were as follows: 277 patterns (CMRSA2 and related); 126 (CMRSA10 and related); 39 (CMRSA7 and related); and 357 (not assigned). Of the CMRSA10, CMRSA2, and CMRSA7 strains, 60·4% (2853/4723), 0% (0/2488), and 74·3% (418/562) of isolates had an indistinguishable PFGE pattern from the respective CMRSA prototype. Of the related PFGE patterns associated with CMRSA10, CMRSA2, and CMRSA7, 36% and 39% of CMRSA10 patterns; 29% and 27% of CMRSA2 patterns; and 23% and 31% of CMRSA7 patterns, were introduced in year 2 and year 3 of surveillance, respectively.
In the Calgary and Edmonton areas, CMRSA10 represented 55·0% and 64·4% of all MRSA isolates, respectively. However, the highest proportion of CMRSA10 was observed in the Northern Alberta Region with 67·7%. In contrast, CMRSA2 was the predominant strain in Central Alberta and Southern Alberta regions (56·9% and 53·3%, respectively). The highest proportion of CMRSA7 was observed in the Southern Alberta Region with 10·9%.
Table 2 shows the patient location at time of clinical specimen collection and the anatomical site of infection. A total of 4859 isolates (54·5%) was obtained in the outpatient setting with 64·2% being CMRSA10. In contrast, 3335 clinical isolates (37·4%) were obtained from hospitalized patients and residents from long-term care facilities where CMRSA2 was the most common strain type isolated with a frequency of 47·6%. CMRSA10 was the most common (88·8%) from correctional facilities. MRSA was most frequently isolated from a skin and soft-tissue source (6749, 75·7% of clinical isolates), of which 4281 (63·4%) were typed as CMRSA10. Urine was the next most common source (679, 7·6% of isolates) of which 537 (79·1%) were CMRSA2. Infections caused by the latter were more likely to be invasive (blood and sterile sites) than CMRSA10-associated infections (P<0·05).
Table 2. Location of clinical specimen collection and source of clinical isolates associated with CMRSA10 and CMRSA2 strain types from July 2005 to June 2008
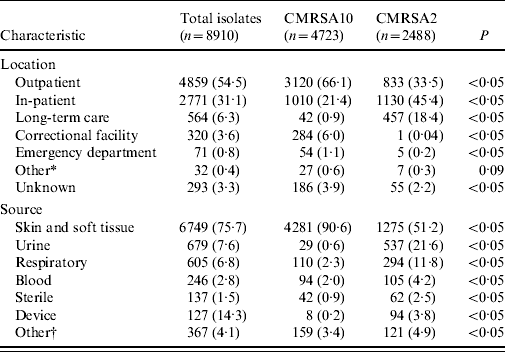
Data represented as no. (%) of isolates.
* Other includes military facility and drop-in centres.
† Other includes drainage from unspecified source or mucous membrane.
Antimicrobial susceptibility
The results of antimicrobial susceptibility testing on blood culture isolates are shown in Table 3. Overall, CMRSA10 and 7 were more susceptible to antimicrobial agents compared to CMRSA2. Representative CMRSA10 non-blood isolates (n=357) from the different regions/areas were compared to CMRSA10 blood isolates (n=63), and there were no significant differences in the antimicrobial susceptibility (data not shown). No resistance to vancomycin, linezolid, rifampin, and quinupristin–dalfopristine was observed in all isolates tested.
Table 3. Antimicrobial resistance profiles of methicillin-resistant Staphylococcus aureus (MRSA) strains isolated from blood
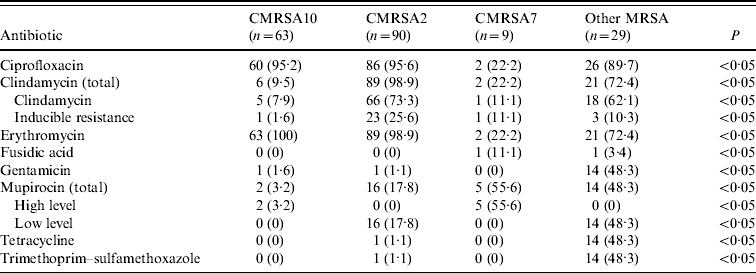
Data represented as no. (%) of resistant isolates.
Discussion
In Canada, there has been a shift in the epidemic strain types over the last several decades, the most recent being the dissemination of CMRSA10 [Reference Cimolai22]. Following a CMRSA10 outbreak in 2004 [Reference Gilbert16], we conducted a prospective microbiological surveillance of all MRSA infections in the province of Alberta in an attempt to describe trends in the epidemiology and implications for clinical management. The calculated annual incidence of all MRSA infection was 107·3 cases/100 000 population in 2008 compared with 57·0 cases/100 000 population in 2005, reflecting a near twofold increase over the study period. This trend was most attributable to the rise of CMRSA10, the predominant strain in Alberta representing >50% of all MRSA infections. The calculated annual incidence of CMRSA10 increased from 21·9 cases/100 000 population in 2005 to 61·5 cases/100 000 population in 2008. Although a direct comparison with other population-based surveillance studies is difficult to make due to different time intervals and definitions used [Reference Klevens14, Reference Fridkin23, Reference Liu24], this striking emergence of CMRSA10 (USA300) has been similarly noted [Reference Huang25, Reference Patel26].
Consistent with other studies [Reference Klevens14, Reference Liu24], males were more likely to acquire MRSA infections. The highest incidence of CMRSA10 infection occurred in the younger 35–44 years age group (76·3 cases/100 000 population), while the highest incidence of CMRSA2 occurred in the >75 years age group. In this older age group, there was also a trend toward a higher incidence of infection with other MRSA strain types, which consisted mainly of CMRSA6 (another HA-MRSA strain) (Fig. 4). This observation further confirms the high prevalence of HA-MRSA infection in the elderly. Blood isolates were more likely to be associated with CMRSA2 than CMRSA10 (P<0·05), probably due to a higher proportion of hospitalized patients with older age and medical comorbidities. Our findings are similar to earlier studies where CA-MRSA strains tend to infect younger patients with fewer medical comorbidities [Reference Moran12, Reference Liu24, Reference Naimi27], and HA-MRSA strains are more likely to infect older patients with prolonged hospitalizations and antibiotic exposures [Reference Simor15, Reference Thompson, Cabezudo and Wenzel28, Reference Monnet29].
Interestingly, geographic variability in the CMRSA distribution was seen with a greater proportion of CMRSA10 (58·4%) in the two major urban centres (Calgary and Edmonton) compared to the rest of the province (40·6%). This may reflect a larger proportion of marginalized population in urban centres [Reference Gilbert16, Reference Simmonds30]. The exception was the Northern Alberta Region, which had the highest proportion of CMRSA10 (67·7%). This is a community that is characterized by rapid industrial development and a young transient population consisting of primarily males, many of whom have migrated from the urban centres for employment. That a significantly higher proportion of CMRSA10 isolates was obtained in the outpatient setting (including those in the emergency department) and accounted for the majority of all skin and soft-tissue source clinical isolates, is consistent with the recent observation that USA300 is the predominant cause of skin and soft-tissue infections in the community [Reference Moran12, Reference King13].
In Alberta, MRSA still remains susceptible to several important anti-MRSA agents. Tetracyclines and trimethoprim–sulfamethoxazole may be considered as empiric therapy for non-invasive MRSA infections. Although more HA-MRSA isolates were resistant to clindamycin, it remains a reasonable therapeutic option for non-invasive CA-MRSA infections. In contrast to other studies done earlier [Reference Liu24, Reference Naimi27], there was a much higher rate of resistance to ciprofloxacin and erythromycin in our data, rendering them ineffective treatments for all MRSA infections. These rates certainly reflect the frequent use of fluroquinolones and macrolides exerting selective pressure in the development of antimicrobial resistance. Interestingly, a low rate of high-level mupirocin resistance was observed in CMRSA10, while the majority of CMRSA7 isolates had high-level mupirocin resistance. An earlier report by Simor et al. described similar trends in mupirocin resistance in MRSA isolates in Canadian hospitals, although the surveillance data was collected prior to the rapid emergence and spread of CMRSA10 in 2004 [Reference Simor31]. In Alberta, mupirocin may be a useful part of a decolonization bundle, especially in targeting high-risk groups and outbreaks.
There has been a dramatic increase of CMRSA10 occurring both in the community and healthcare settings in Alberta. Although the public and the medical profession are becoming more aware of this trend, the sudden surge cannot be attributed to increased vigilance in surveillance alone. Reasons for this increase are not entirely clear. In Alberta, 60% of CMRSA10 strains have a PFGE pattern indistinguishable from that of the prototype CMRSA10 strain. Although direct transmission among those in a closed population, such as the marginalized and homeless, may account for high rates of strains having an indistinguishable PFGE fingerprint, it is possible that this CMRSA10 strain is able to disseminate more readily than other strains. The antimicrobial susceptibility profiles of CMRSA10 blood isolates have been similar over the 3 years, suggesting minimal introduction of novel resistance determinants into these strains. Moreover, the development of new but related CMRSA10 strains by PFGE has been stable from year to year, suggesting that no major changes in the genetic make-up of the strains have occurred. Ongoing surveillance of CMRSA10 will be important to determine any changes in virulence or antimicrobial resistance. Reasons for a certain clonal dominance and the mechanism of its evolution have been the subject of much debate and speculation [Reference Deurenberg and Stobberingh32, Reference DeLeo and Chambers33].
The rates of MRSA are projected to stabilize in Alberta with greater efforts in province-wide surveillance and infection prevention and control policies having a certain impact. Stabilization of MRSA rates has been observed in other countries such as France and the UK [3]. Our findings have several important implications. Given that CMRSA10 constitutes the majority of MRSA in Alberta, prevention and control measures should encompass more than just healthcare facilities. From our current understanding of risk factors associated with CA-MRSA, greater emphasis on personal hygiene and environmental control is warranted in specialized institutions such as correctional facilities and homeless shelters. In addition, control measures need to be targeted at the community level. Education regarding transmission and containment of MRSA at the community level has been identified as a gap in the control of CA-MRSA [Reference Hawkes34]. This trend also has relevance in clinical management of infections. For instance, knowing that CMRSA10 is the predominant clone in MRSA skin and soft-tissue infections will allow clinicians to use targeted antimicrobial therapy after an assessment of certain risk factors. However, ongoing surveillance is needed to examine the trends in strain distribution, which may have implications for change in antimicrobial susceptibility.
There are several limitations in our study. Our data only captured those patients who had cultures obtained, underestimating the true burden of disease where cultures were not routinely done. Because only the first clinical specimen from each patient was used to determine the site of infection, other potentially more relevant specimens from the same patient could have been excluded in the analysis. Moreover, a propensity for certain MRSA strains to cause recurrent infections could not be analysed. However, one of the main objectives of the study was to describe the change in incidence of MRSA rather than rate of recurrence, which required adopting formal criteria for determining incidence. Similar criteria have been employed elsewhere [Reference Liu24]. Finally, we were not able to capture detailed patient risk factors, comorbidities, and timing of hospitalizations.
The strength of this descriptive epidemiological study lies in its comprehensive molecular-typing analysis of all MRSA strains isolated within the entire population of Alberta over a 3-year period and the ability to track the geographical spread of specific MRSA strains within the province. Molecular typing of MRSA as it was used in our study may become increasingly important because the distinction between healthcare-associated and community-associated strains may be blurring as CMRSA10 migrates into healthcare settings and becomes the predominant epidemic type [Reference Klevens35–Reference Patel37]. However, more commonly used epidemiological designations are still clinically relevant since molecular analysis cannot be routinely employed.
In Alberta, CMRSA10 has now become the predominant epidemic type. The changing epidemiology of MRSA highlights the importance of ongoing regional surveillance. As we have observed in our province, it not only impacts infection control measures and antimicrobial utilization, but also public health initiatives [Reference Allen36]. Research into transmission, risk factors, and infection control strategies such as screening and decolonization, is needed. Further genetic characterization and analysis of MRSA with regards to SCCmec, virulence profiling, and detection of resistant determinants will provide a better understanding of the evolution of these strains over time.
ACKNOWLEDGEMENTS
We thank Vincent Li (ProvLab) for assistance with background work in PFGE typing analysis. We acknowledge the contributions of Alberta MicroNet in providing support for the surveillance programme and submission of MRSA isolates to ProvLab. Alberta MicroNet include the following laboratories in Alberta: Banff Mineral Springs Hospital Laboratory, Banff; Bonnyville Health Centre Laboratory, Bonnyville; Calgary Laboratory Services, Calgary; Chinook Health Region Main Laboratory, Lethbridge; DynaLIFEDX Diagnostic Laboratory, Edmonton; High River Hospital Laboratory, High River; Medicine Hat Diagnostic Laboratory, Medicine Hat Regional Hospital Laboratory, Medicine Hat; Northern Lights Regional Health Centre, Ft. McMurray; Queen Elizabeth II Hospital Laboratory, Grand Prairie; Red Deer Regional Hospital Laboratory, Red Deer; St. Mary's Hospital Laboratory, Camrose; University of Alberta Hospital Laboratory, Edmonton; Wainwright Health Centre Laboratory, Wainwright; and Westlock Healthcare Centre Laboratory, Westlock.
DECLARATION OF INTEREST
None.









