INTRODUCTION
Anaplasmataceae agents are obligate intracellular bacteria, resident in phagosomes and belonging to the order Rickettsiales, family Anaplasmataceae, α sub-division of Proteobacteria, whose cycle in the environment involves complex interactions between invertebrate vectors and vertebrate hosts. Some Anaplasmataceae agents, such as Ehrlichia spp. and Anaplasma spp., have great importance in veterinary and human medicine. For instance, while Ehrlichia canis is the etiological agent of canine monocytic ehrlichiosis (CME), a widespread tick-borne disease among dogs around the world, A. phagocytophilum and Ehrlichia chaffeensis are the main agents responsible for human granulocytic anaplasmosis and human monocytic ehrlichiosis (HME), respectively, in humans in North Hemisphere. The clinical presentation caused by Anaplasmataceae agents includes fever, headache, petechiae, myalgia, althralgia, rash, cough, nausea, vomiting, abdominal pain, diarrhea, leukopenia, thrombocythopenia and elevated liver enzymes [Reference Dumler1].
In Brazil, the recent molecular detection of A. phagocytophilum [Reference Santos2, Reference Silveira3] in domestic dogs, associated with the detection of new genotypes of Ehrlichia spp. and Anaplasma spp. in deer [Reference Machado4, Reference Mongruel5], birds [Reference Machado6], wild canids [Reference André7], domestic and wild felids [Reference André8, Reference André9], and rodents [Reference Benevenute10] has shown a diversity of Anaplasmataceae agents circulating in domestic and wild mammals.
The aim of the present study was to investigate the occurence of Anaplasmataceae agents (Ehrlichia and Anaplasma) in wild mammals, domestic dogs and their respective ectoparasites in the Pantanal wetland, in order to shed some light on the role of wild animals in the epidemiology of Anaplasmataceae agents in wild environments. The Pantanal is a 160 000 km2 floodplain located in the centre of the South American continent. It is a mosaic of seasonally inundated native grasslands, savannas and scrub savannas, river corridors, lakes, gallery forests and patches of scrub and semi-deciduous forests. In this highly diverse environment, in which humans, wild and domestic animals and arthropods species share the same habitat, the landscape dynamics and resource availability change according to a multi-year variation of flooding intensity [Reference Herrera11]. Although the number of studies investigating the role of wild animals in the epidemiology of diseases caused by Anaplasmataceae agents has increased around the world, few studies [Reference Melo12] have focused on the occurrence of these agents in the Pantanal wetland.
MATERIAL AND METHODS
Ethical aspects
All animal captures were in accordance with the licenses obtained from the Brazilian Government Institute for Wildlife and Natural Resources Care (IBAMA) (license numbers 38145 and 38787-2) and were endorsed by the Ethics Committee of FCAV/UNESP University (Faculdade de Ciências Agrárias e Veterinárias, Universidade Estadual Paulista ‘Júlio de Mesquita Filho’, Câmpus Jaboticabal) no. 006772/13.
Study area
The fieldwork was conducted at the Nhumirim ranch (56°39′ W, 18°59′S), located in the central region of the Pantanal, municipality of Corumbá, state of Mato Grosso do Sul, central-western Brazil (Fig. 1). This region is characterized by a mosaic of semi-deciduous forest, arboreal savannas, seasonally flooded fields covered by grasslands with dispersed shrubs and several temporary and permanent ponds. The Pantanal is the largest Neotropical floodplain, being well known for its rich biodiversity. Two well-defined seasons are recognized in that region: a rainy summer (October–March) and a dry winter (April–September) [Reference Sousa13, Reference Sousa14].
Fig. 1. Capture sites. Map of Mato Grosso do Sul State, central-western Brazil, showing the Pantanal region, where animals samples were sampled in the present study.
Animals sampled
Between the years of 2013 and 2015, four field expeditions were performed in August 2013, October 2013, August 2014 and March 2015. Free-ranging C. thous (crab-eating fox), N. nasua (coati) and L. pardalis (ocelot) were caught using a Zootech® (Curitiba, PR, Brazil) model wire box live trap (1 × 0·40 × 0·50 m3), which was made with galvanized wire mesh and baited with a piece of bacon every afternoon. Twenty traps were placed on the ground at 2 km intervals, left open during 24 h and checked twice a day for 12 days. The animals were immobilized with an intramuscular injection of zolazepan and tiletamine at dosages of 8 mg/kg for ocelots and 10 mg/kg crab-eating foxes and coatis. Blood samples were collected by puncture of the cephalic vein stored in Vacutainer® containing EDTA and stored at −20 °C until DNA extraction.
Small mammals (rodents and marsupials) were captured using live traps (Sherman® – H. B. Sherman Traps, Tallahassee, FL, USA and Tomahawk® Tomahawk Live Traps, Tomahawk, WI, USA) baited with a mixture of banana, peanut butter, oat and sardines. Traps were set up for seven consecutive nights along linear transects, placed on the ground at 10 m intervals and alternating between trap type in two field expeditions (August 2014 and March 2015). The total capture effort was 200 traps-nights, equally distributed in two expeditions (August 2014 and March 2015). The captured rodents and marsupials were euthanized in order to perform the identification of the animal's species, based on external and cranial morphological characters and karyological analyses. The animals were firstly anesthetized with an intramuscular injection of ketamine (10–30 mg/kg) with acepromazine (5–10 mg/kg) for rodents (proportion 9 : 1), or xylazine (2 mg/kg) for marsupials (1 : 1). After anesthesia, the animals were euthanized with potassium chloride, which doses ranged from 75 to 150 mg/kg. Spleen samples were collected and stored in absolute alcohol (Merck®, Kenilworth, Nova Jersey, USA).
A total of 256 animals were captured, including 158 carnivores, including 78 C. thous, 31 N. nasua, seven L. pardalis, 110 wild rodents (77 Thrichomys fosteri, 25 Oecomys mamorae and eight Clyomys laticeps) and 30 wild marsupials (14 Thylamys macrurus, 11 Gracilinanus agilis, four Monodelphis domestica and one Didelphis albiventris). Additionally, 42 blood samples from domestic dogs cohabiting the same studied area were collected [Reference Sousa13, Reference Sousa14].
Ectoparasites identification
Ticks and fleas found parasitizing the sampled animals were detected by inspection of the skin and carefully removed by forceps or manually. The identification was performed using a stereomicroscope (Leica® MZ 16A, Wetzlar, Germany) and following taxonomic literature for adult tick genera [Reference Guimarães15, Reference Martins16], and Amblyomma spp. nymphs [Reference Martins17]. Amblyomma spp. larvae could not be identified to the species level considering the insufficient literature available until now. The identification of fleas was performed following previously described taxonomic keys [Reference Linardi and Guimarães18].
Serological assays
Canids, felids, coatis and rodents’ serum samples were individually tested by Indirect Fluorescent Antibody Test (IFAT) in order to detect IgG antibodies to E. canis and A. phagocytophilum. For that purpose, E. canis and A. phagocytophilum crude antigens were cultivated in DH82 cells and HL-60 cells infected with Jaboticabal strain of E. canis strain [Reference Aguiar19] and Webster strain of A. phagocytophilum, respectively. Briefly, antigen slides were removed from storage and allowed to thaw at room temperature for 30 min. Ten microliters of twofold dilutions of sera (starting at 1 : 80, the cut-off for E. canis and A. phagocytophilum) were placed in wells on antigen slides. On each slide, previously determined non-reactive and reactive serum samples to E. canis were used as negative and positive controls, respectively [Reference André9, Reference Sousa20]. A known positive A. phagocytophilum serum (titer 1 : 2560) was obtained from a horse experimentally infected with the Webster strain of A. phagocytophilum at Department of Veterinary Pathology, UNESP, Jaboticabal, São Paulo, Brazil. A horse serum sample negative for A. phagocytophilum was used as negative control. Slides were incubated at 37 °C in a moist chamber for 30 min, washed three times in PBS (pH 7·2) for 5 min, and air dried at room temperature. Then, the slides were incubated with fluorescein isothiocyanate labeled anti-mouse IgG (Sigma®, St. Louis, USA) for wild rodents, goat anti-dog IgG (Sigma®, St. Louis, USA) for C. thous and domestic dogs, goat anti-cat IgG (Sigma®, St. Louis, USA) for L. pardalis, goat anti-raccoon IgG (Sigma®, St. Louis, USA) for N. nasua and goat anti-horse IgG (Sigma®, St. Louis, USA) for A. phagocytophilum control serum sample. Anti-cat conjugate (dilution of 1 : 32) for feline samples, anti-dog conjugate for the canine samples (dilution of 1 : 32), anti-raccoon (dilution of 1 : 10), anti-mouse conjugate for the wild rodents samples and anti-horse (dilution of 1 : 64) were diluted according to the manufacturer's instructions and then added to each well. These slides were incubated again at 37 °C, washed three times in PBS, once more in distilled water, and air dried at room temperature. Finally, slides were overlaid with buffered glycerin (pH 8·7), covered with glass coverslips, and examined with a fluorescence microscope (Olympus®, Tokyo, Japan). Unfortunately, it was not possible to test the marsupial's serum samples, due to the unavailability of conjugate for this animal species in the laboratory.
Molecular assays
DNA was extracted from 200 µl of each whole blood (158 wild carnivores and 42 domestic dogs) and 10 mg of spleen (140 small mammals) samples using the DNeasy Blood & Tissue Kit (QIAGEN®, Valencia, CA, USA), according to the manufacturer's instructions. DNA was extracted from 523 tick samples, including 228 (43·5%) adults, 256 (48·9%) pooled nymphs, and 39 (7·4%) pooled larvae. DNA extraction from ticks was processed in pools for nymphs (up to five individuals) and larvae (up to 10 individuals). The adults were processed individually. A total of 39 pooled fleas samples (each one consisting of up to five individuals) were used for DNA extraction. Ticks and fleas were macerated and DNA extracted using the same kit above mentioned [Reference Sousa13, Reference Sousa14].
In order to verify the presence of amplifiable DNA in the samples, internal control polymerase chain reaction (PCR) assays targeting fragments of mammalian glyceraldehyde-3-phosphate dehydrogenase (GAPDH) [Reference Birkenheuer, Levy and Breitschwerdt21], tick mitochondrial 16S rRNA genes [Reference Black and Piesman22] and flea cytochrome-c oxidase subunit I (COX1) [Reference Folmer23] genes were performed (Table S1).
First, a previously described broad range multiplex qPCR protocol based on groEL gene (Table S2) to detect and quantify Ehrlichia spp. and Anaplasma spp. [Reference Benevenute10] and cPCR assays targeting 16SrRNA genes [Reference Murphy24, Reference Massung25] (Table S1) of Ehrlichia spp. and Anaplasma spp. were performed. The limit of detection of groE gene fragment quantification for Ehrlichia spp. and Anaplasma was 10 copies/μl [Reference Benevenute10]. All positive samples in groEL qPCR and 16SrRNA gene cPCR reactions were tested using previously described specific qPCR assays to detect and quantify A. phagocytophilum (msp2) [Reference Drazenovich, Foley and Brown26], E. canis (dsb) [Reference Doyle27] and E. chaffeensis (vlpt) [Reference Reller28] (Table S2). Additionally, cPCR assays targeting four other protein-coding genes, namely groESL [Reference Sumner, Nicholson and Massung29, Reference Nicholson30] for Ehrlichia and Anaplasma spp., omp1 [Reference Inayoshi31] for Ehrlichia spp., dsb [Reference Doyle27] for E. canis, trp36 [Reference Aguiar32] for E. canis and Ehrlichia minasensis sp. nov., and msp5 [Reference Singh33] for Anaplasma spp. (Table S1).
The TaqMan qPCR reactions based on groEL, msp2, dsb and vlpt genes were performed with a final volume of 10 µl containing 5 µl GoTaq® Probe qPCR Master Mix (Promega Corporation, Madison USA), 1·2 µM of each primer and hydrolysis probe (Table S2) and 1 µl of each DNA sample. PCR amplifications were performed in low-profile multiplate unskirted PCR plates (BioRad®, CA USA) using a CFX96 Thermal Cycler (BioRad®, CA USA). Serial dilutions were performed aiming to construct standard curves with different plasmid DNA concentrations (Integrated DNA Technologies®, Coralville, Iowa, USA) (2·0 × 107–2·0 × 100 copies/μl). The number of plasmid copies was determined in accordance with the formula (ng/μl DNA/plasmid size (bp) × 660]) × 6·022 × 1023 × plasmid copies/μl. Each qPCR assay was performed including duplicates of each DNA sample. All the duplicates showing Cq difference values higher than 0·5 were re-tested. Amplification efficiency (E) was calculated from the slope of the standard curve in each run using the following formula (E = 10–1/slope). To determine the limit of detection from the qPCR assay, the standard curves generated by 10-fold dilutions were used to determine the amount of DNA that could be detected with 95% of sensitivity [Reference Bustin34].
The cPCR reactions contained 10X PCR buffer (Life Technologies®, Carlsbad, CA, USA), 1 mM MgCl2 (Life Technologies®, Carlsbad, CA, USA), 0·2 mM deoxynucleotide triphosphate (dNTPs) mixture (Life Technologies®, Carlsbad, CA, USA), 1·5 U Taq DNA Polymerase (Life Technologies®, Carlsbad, CA, USA), and 0·5 µM of each primer (Integrated DNA Technologies®, Coralville, IA, USA) (Table S1). Ehrlichia canis and Anaplasma spp. DNA positive controls were obtained from naturally infected cats [Reference André9]. Ultra-pure sterile water (Life Technologies®, Carlsbad, CA, USA) was used as negative control in all PCR assays. PCR products were separated by electrophoresis on a 1% agarose gel stained with ethidium bromide (Life Technologies®, Carlsbad, CA, USA). In order to prevent PCR contamination, DNA extraction, reaction setup, PCR amplification and electrophoresis were performed in separated rooms. The gels were imaged under ultraviolet light using the Image Lab Software version 4.1 (Bio-Rad®). The reaction products were purified using the Silica Bead DNA gel extraction kit (Thermo Fisher Scientific®, Waltham, MA, USA). Sanger sequencing was performed for all positive samples in cPCRs assays, using the BigDye® Terminator v3.1Cycle Sequencing Kit (Thermo Fisher Scientific®, Waltham, MA, USA) and ABI PRISM 310DNA Analyzer (Applied Biosystems®, Foster City, CA, EUA).
Bioinformatics/phylogenetic analysis
Sequences obtained from positive samples in cPCR assays were first submitted to a screening test using Phred-Phrap software version 23 [Reference Ewing and Green35] in order to evaluate the electropherogram quality and to obtain consensus sequences from the alignment of sense and antisense sequences. The BLAST program [Reference Altschul36] was used to analyze the sequences of nucleotides (BLASTn), aiming to browse and compare with sequences previously deposited in an international database (GenBank) [Reference Benson37]. All sequences that showed appropriate quality standards and identity with Ehrlichia spp. or Anaplasma spp. were deposited in Genbank. The sequences were aligned with sequences published in GenBank using MAFFT software, version 7 [Reference Katoh and Standley38].
Phylogenetic inference was based on Bayesian Inference (BI) method. The BI analysis was performed with MrBayes 3.1.2 [Reference Ronquist and Huelsenbeck39]. Markov chain Monte Carlo (MCMC) simulations were run for 109 generations with a sampling frequency of every 100 generations and a burn-in of 25%. The best model of evolution was selected by the program jModelTest2 (version 2.1.6) on XSEDE [Reference Darriba40], under the Akaike Information Criterion (AIC) [Reference Miller, Pfeiffer and Schwartz41]. All phylogenetic analyses were performed using CIPRES Science Gateway [Reference Miller, Pfeiffer and Schwartz41]. The trees were examined in Treegraph 2.0.56-381 beta [Reference Stover and Muller42].
RESULTS
A total of 1582 ticks parasitizing the sampled mammals were collected, of which 1033 (65·2% [115 adults and 918 nymphs]) belonging to Amblyomma sculptum Barlese, 1888 species; 241 (15·2% [78 adults and 163 nymphs]) belonging to Amblyomma parvum Aragão, 1908 species; 32 (2%) Amblyomma ovale (Koch, 1844) adults; one (0·06%) Amblyomma tigrinum Koch, 1844 adult; one (0·06%) Rhipicephalus (Boophilus) microplus (Canestrini, 1887) adult; one (0·06%) Rhipicephalus sanguineus sensu lato adult; four (0·2%) Amblyomma auricularium (Conil, 1878) nymphs; and 269 (17%) Amblyomma larvae. Besides, a total of 80 Polygenis (Polygenis) bohlsi bohlsi fleas were also collected (Table 1) [Reference Sousa13, Reference Sousa14].
Table 1. Tick species collected from wild mammals captured between August 2013 and March 2015 in Pantanal wetland, Brazil

L, larvae; N, nymphs; M, male adults; F, female adults; Noanim, number of sampled animals; No infest, number of infested animals according to host species.
a A. sculptum, Amblyomma sculptum; A. parvum, Amblyomma parvum; A. tigrinum, Amblyomma tigrinum; A. ovale, Amblyomma ovale; A. auricularium, Amblyomma auricularium; R.(B.) microplus, Rhipicephalus (Boophilus) microplus; R. sanguineus s.l., Rhipicephalus sanguineus sensu lato.
All 298 DNA animal samples amplified the predicted product for GAPDH gene. Out of 523 DNA sampled ticks, 31 [5·9% (23 A. parvum adults, four A. sculptum adults, one A. ovale adult, one A. parvum nymph and two pooled Amblyomma spp. Larvae)] showed negative results for the tick mitochondrial 16S rRNA gene and were excluded from subsequent analyses. Only one flea DNA sample did not amplify the predicted product for COX1 gene and was also excluded from subsequent analyses [Reference Sousa13, Reference Sousa14].
Overall, 14 (17·9%) C. thous, seven (16·6%) C. familiaris and one(3·2%) N. nasua were seroreactive (titre⩾80) to E. canis. The seroreactive animals showed titres for E. canis ranging from 80 to 1280. No wild rodent showed to be seroreactive to E. canis antigen. Ticks were collected from 12 (54·5%) of 22 seroreactive animals. None of the animals was seroreactive for A. phagocytophilum (Table 2).
Table 2. Number of domestic dogs and wild mammals seroreactive to Ehrlichia canis in Pantanal wetland, Brazil

Twenty-seven animals (9%) were positive in cPCR assays for Ehrlichia spp. based on 16SrRNA gene, including nine (21·4%) C. familiaris, two (7·6%) C. thous, one (3·8%) N. nasua, four (15·3%) T. fosteri, four (15·3%) O. mamorae, three (11·5%) T. macrurus, two M. domestica (50%) and two (7·6%) G. agilis. Additionally, eight A. sculptum adults, four A. parvum adults, 13 A. sculptum nymphal pools, two Amblyomma larvae pools and one (2·5%) Polygenis (P.) bohlsi bohlsi flea pool showed positive results in cPCR assays for Ehrlichia spp. based on 16SrRNA gene. Two A. sculptum adults positive in cPCR assays for Ehrlichia spp. based on 16SrRNA were collected from a C. thous that was also positive in Ehrlichia-cPCR assay based on 16SrRNA gene. Four (9·5%) dogs were positive in qPCR assays for Ehrlichia spp. based on groEL gene. Nine dogs (21·4%) and one Polygenis (P.) bohlsi bohlsi flea pool also showed positive results in a specific qPCR for E. canis based on dsb gene (Table 3). All 16S rRNA gene-Ehrlichia sequences were deposited in Genbank international database under the accession numbers KY499155-KY499181 and KY930380-KY930407.
Table 3. Number of domestic dogs and wild mammals positive in qPCR and cPCR assays for Ehrlichia and Anaplasma

Seven (22·5%) N. nasua, one (1·2%) C. thous, one (14·2%) L. pardalis, two (2·5%) T. fosteri, one (12·5%) C. laticeps, one (9%) G. agilis, one (7·1%) T. macrurus, two (4·7%) C. familiaris, (0·9%), two A. sculptum nymphal pools and one (3·1%) A. ovale adult showed positivity in cPCR assays for Anaplasma spp. based on 16SrRNA gene. Two (3·5%) T. fosteri, one (4%) O. mamorae, two (18·1%) G. agilis, one (7·1%) T. macrurus, 10 (3·1%) A. sculptum adults, five (2·4%) A. sculptum nymphal pools, two (1·5%) A. parvum adults, two (6·2%) A. ovale adults, and one (2·5%) Amblyomma larvae pool, were positive in qPCR for Anaplasma spp. based on groEL gene (Table 3). Positive samples in cPCR (16SrRNA gene) and qPCR (groEL) assays for Anaplasma spp. did not show positive results in cPCR assays based on groESL and msp5 genes neither in the qPCR assay for A. phagocytophilum based on msp2 gene. All 16SrRNA gene-Anaplasma sequences were deposited in Genbank international database under the accession numbers KY499182 -KY499201.
Co-positivity was observed between the molecular assays to Ehrlichia spp. and Anaplasma spp. or between the molecular and serological assays to Ehrlichia spp. and Ehrlichia canis in two dogs, one N. nasua, one C. thous, one T. macrurus and one G. agilis (Table 4).
Table 4. Co-positivity observed between the molecular assays to Ehrlichia spp. and Anaplasma spp. and between molecular and serological assays to Ehrlichia spp. and Ehrlichia canis

The BLAST analysis of the 16SrRNA gene of Ehrlichia spp. showed that sequences obtained from C. thous, N. nasua, M. domestica, T. macrurus, G. agilis, T. fosteri, O. mamorae, dogs, a pool of Polygenis (P.) bohlsi bohlsi, A. sculptum nymphal pools, A. sculptum adults, Amblyomma spp. larvae pools and A. parvum adults samples, showed 99%–100% identity with Ehrlichia spp. sequences obtained from free-living wild animals from Brazil (KX898136, JQ260861, JQ260855), an E. canis sequence from Brazil (JX118827), an E. canis sequence obtained from R. sanguineus ticks collected in Philippines (JN121379), and with an E. canis sequence from Malaysia (KR920044). The 16SrRNA gene sequences of Anaplasma spp. obtained from N. nasua, L. pardalis, C. thous, dogs, T. fosteri, C. laticeps, T. macrurus, G. agilis, A. sculptum nymphal pool and A. ovale samples, showed 98–100% of identity with an Anaplasma spp. sequence obtained from Amblyomma cajennense sensu lato ticks (KJ831219) collected in the state of Mato Grosso, Brazil, Anaplasma platys (KU500914, KU534873) and A. phagocytophilum (CP006618, GU064900) by BLAST analysis (Table S3).
The phylogenetic analysis based on 16SrRNA gene fragments of Ehrlichia spp. grouped the sequences obtained from mammals, ticks and a flea pool (KY499155-KY499181 and KY930380-KY930407) together and in the same branch of E. canis sequences from Brazil (EF195135) and other countries (EF011111, EU106856, U26740) and with Ehrlichia spp. sequences obtained from free-living N. jubata and wild felids from Brazil (KX898136, EU376114), based on Bayesian analysis, with clade support of 61 (Fig. 2). Regarding the phylogenetic analysis of Anaplasma based on 16SrRNA gene fragments, Anaplasma spp. sequences obtained from one N. nasua (KY499193), one C. familiaris (KY499188) and one A. sculptum nymphal pool (KY499182) were grouped in the same clade and together with one sequence obtained from Carcara plancus from Brazil (JN217096), one A. phagocytophilum sequence (GU236670) obtained from an Austrian dog and one Anaplasma sp. sequence (KF964051) obtained from a domestic cat from Brazil, based on BI analysis. Four Anaplasma spp. 16SrRNA gene sequences obtained from N. nasua (KY499184, KY499187, KY499194, KY499195), one sequence obtained from L. pardalis (KY499183), one sequence obtained from C. thous (KY499185), two sequences obtained from T. fosteri (KY499197, KY499198), one sequence obtained from C. laticeps (KY499196), one sequence obtained from T. macrurus (KY499200), one sequence obtained from G. agilis (KY499199), one sequence obtained from one A. sculptum nymphal pool (KY499201) and one sequence obtained from one A. ovale adult (KY499191) were positioned in the same clade of A. bovis (LC012812), with clade support of 84 (Fig. 3).
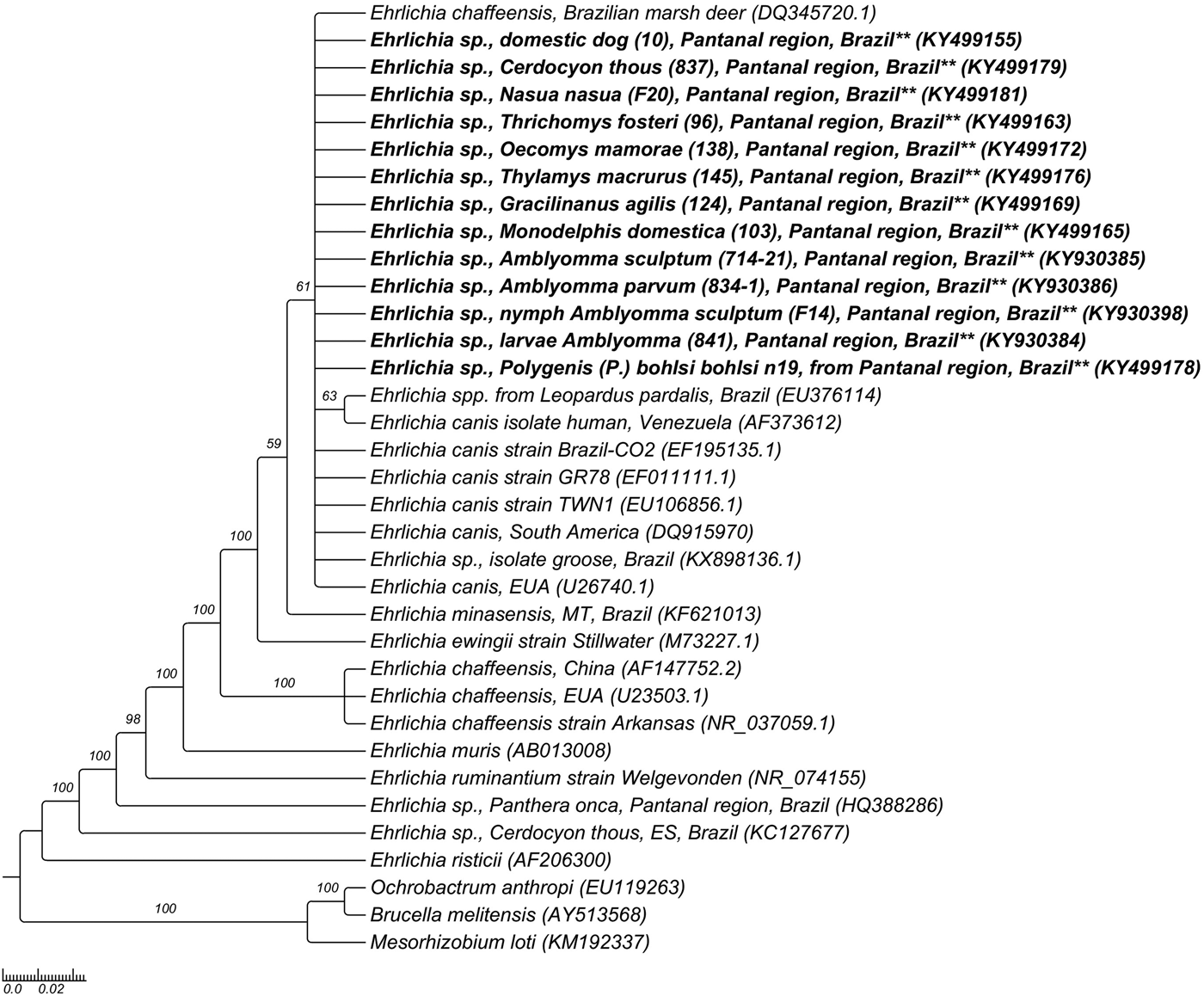
Fig. 2. Phylogenetic tree constructed with 860 pb Ehrlichia spp.-16SrRNA sequences, using Bayesian method and GTR + G + I evolutionary model. Numbers at nodes correspond to Bayesian posterior probabilities over 50, using Mesorhizobium loti (KM192337), Brucella melitensis (AY513568) and Ochrobactrum anthropi (EU119263) as outgroups.

Fig. 3. Phylogenetic tree constructed with 600 pb Anaplasma spp.-16SrRNA sequences, using Bayesian method and GTR + G + I evolutionary model. Numbers at nodes correspond to Bayesian posterior probabilities over 50, using Mesorhizobium loti (KM192337), Brucella melitensis (AY513568) and Ochrobactrum anthropi (EU119263) as outgroups.
DISCUSSION
In the present study, the circulation of E. canis among dogs in Brazilian Pantanal wetland was confirmed by both molecular and serological methods. Considering the pathogenic potential of E. canis in domestic dogs [Reference Dumler1], the role of these animals as a source of E. canis infection to wild carnivores cohabiting the same area and the impact of the infection on wildlife health requires further studies.
Reports on seropositivity to E. canis among wild carnivores are scarce in Brazil. In fact, there is only one report concerning the serological detection of antibodies to E. canis among wild felids maintained in captivity in zoos in the states of São Paulo and Mato Grosso state, Brazil [Reference André8]. Herein, seroprevalence rates of 17·9% and 3·2% were found among free-living C. thous and N. nasua, respectively. However, seropositive animals showed negative results in the specific qPCR assay for E. canis based on dsb gene. Due to the unavailability of other Ehrlichia species antigens, serological cross-reactions between E. canis and closely related agents could not be excluded
Ehrlichia spp. DNA was detected in two C. thous from Pantanal wetland. Previous studies had already detected Ehrlichia spp. DNA among wild canids (C. thous, Speothos venaticus and Canis lupus) maintained in captivity in Brazilian zoos [Reference André8] and among road-killed C. thous in the state of Espírito Santo, southeastern Brazil [Reference Almeida43]. However, it is worth noticing that while Ehrlichia spp. DNA has been detected among Brazilian canids, no clinical signs suggestive of ehrlichiosis have been reported until now in this group of wild mammals. Future studies are needed in order to clarify the role of these animals in the epidemiology of Ehrlichia spp. in nature.
Among the wild felids, all seven L. pardalis trapped in this present study showed negative results in molecular and serological assays for Ehrlichia spp. In a previous study, an Ehrlichia dsb sequence obtained in one out of 10 jaguars also sampled in the Pantanal biome were closely related to Ehrlichia ruminantium [Reference Widmer44]. While A. sculptum (formerly named as A. cajennense), Amblyomma triste and R. (B.) microplus were previously found parasitizing jaguars in Pantanal [Reference Widmer44], only A. parvum was found parasitizing L. pardalis in the present study at the time of sample collection. Further studies are needed to determine the real role of different tick species in the transmission of ehrlichial agents among wild felids in Brazil.
Few studies have assessed the occurrence of Anaplasmataceae agents in marsupials. Previously, antibodies to E. canis were detected in 14·6% of opossums (Didelphis aurita and Didelphis albiventris) trapped in São Paulo state, southeastern Brazil [Reference Melo45]. Although serological assays were not performed in marsupials’ serum samples collected in the present study, Ehrlichia DNA was detected in 23·3% of sampled marsupials. The present study highlights that marsupials may participate in the biological cycles of Ehrlichia in the environment. To the best authors’ knowledge, this was the first molecular evidence of exposure to Ehrlichia spp. among free-living marsupials in the world until now.
Herein, Ehrlichia DNA was detected in 7·2% of wild rodents (T. fosteri and O. mamorae) sampled in southern Pantanal. Recently, Ehrlichia DNA was detected in 24% of wild rodents (O. mamorae, T. fosteri, C. laticeps and Calomys cerqueirai) sampled in the biome Pantanal [Reference Benevenute10]. Although rodents are considered hosts for E. chaffeensis in China and Korea [Reference Chae46–Reference Gao48], the role of wild rodents in natural cycles of Ehrlichia species and the arthropod vectors involved in the transmission routes remain unknown in Brazil.
Ehrlichia canis DNA was detected in one Polygenis (P.) bohlsi bohlsiflea pool collected from a specimen of T. fosteri rodent that was negative in PCR assays for Ehrlichia spp. In a previous study, E. canis DNA was detected in one (1/1) Cediopsylla inaequalis and 3% (2/75) of the Xenopsylla cheopis fleas collected from red foxes (Vulpes vulpes) in Italy [Reference Torina49]. Although E. canis DNA was detected in a Polygenis (P.) bohlsi bohlsiflea pool by qPCR and cPCR assays, we believe that the positivity observed was due to residual ehrlichial DNA from host blood in the siphonapteran digestive tract.
Anaplasma DNA was detected among wild carnivores (C. thous, L. pardalis and N. nasua) and dogs sampled in the present study. Previously, Anaplasma DNA was detected in three wild felids (L. tigrinus) and one wild canid (S. venaticus) maintained in captivity in zoos in the state of São Paulo [Reference André7]. Additionally, wild rodents and marsupials were also positive for Anaplasma spp. Recently, Anaplasma DNA was detected in nine rodents (Rattus rattus, Akodon sp., Sphiggurus villosus and Calomys cerqueirae) sampled in Atlantic Forest and Caatinga biomes in Brazil [Reference Benevenute10]. In the northern region of the Brazilian Pantanal, Anaplasma DNA was also detected in only one Hylaeamys megacephalus wild rodent (1/4) [Reference Wolf50]. Additional studies should be done in order to access the role of wild carnivores, rodents and marsupials in the Anaplasma epidemiology in South America.
The arthropod vectors involved in Anaplasma and Ehrlichia species transmission cycles among wild mammals in Brazil are still unknown. Previously, unclassified Anaplasma spp. and Ehrlichia spp. were detected in one A. sculptum tick collected from a dog [Reference Melo12] and in A. sculptum (formerly named as A. cajennense), A. triste and Amblyomma spp. nymph ticks collected from P. onca [Reference Widmer44], respectively, in Pantanal biome. The present study reports the molecular detection of Anaplasma spp. among A. sculptum, A. ovale and A. parvum ticks, and Ehrlichia spp. in A. sculptum, A. parvum and Amblyomma spp. larvae. According to some authors, questing tick samples are preferable in order to assess the real role of arthropods as vectors of Anaplasmataceae agents [Reference Estrada-Peña51]. Considering the fact that we detected Ehrlichia and Anaplasma DNA in ticks found parasitizing sampled animals, we assumed that tick positivity could be related to the remnant of infected host blood meal.
The phylogenetic inferences based on a small fragment of 16S rRNA gene positioned the Ehrlichia and Anaplasma genotypes in the same clade of E. canis and A. bovis, respectively. The variable amplification of different target genes in the present study, which precluded additional phylogenetic inferences, could be explained by low bacteraemia level in mammalian blood or spleen samples and ectoparasites. Moreover, PCR protocols used for amplification of different target genes have been proven unsuitable for amplification of variants of Anaplasma and Ehrlichia species infecting wild mammals in Brazil, as previously reported [Reference Machado6, Reference André7, Reference Benevenute10]. The phylogenetic analysis based on short 16S rRNA gene fragments did not provide sufficient genetic discrimination to allow the identification of Ehrlichia and Anaplasma species. Although genotypes closely related to E. chaffeensis, E. canis and A. phagocytophilum were detected in deer [Reference Machado4, Reference Mongruel5], wild carnivores [Reference André7], birds [Reference Machado6], and domestic cats [Reference André9] in Brazil, these new genotypes have not been isolated so far. Alternatively, species-specific qPCR assays might not have sufficient sensitivity to detect the presence of E. canis and E. chaffeensis in wild animals’ biological samples in the present study.
In conclusion, the present study revealed that wild animals and ticks in southern Pantanal region, Brazil, are exposed to Anaplasmataceae agents. The role of wild animals in the epidemiology of Anaplasmataceae agents as reservoirs and the impact of the infection on wildlife health should be further investigated. Although Anaplasma and Ehrlichia have been molecularly detected among ticks and fleas collected from sampled animals, the vectors involved in the natural cycles of Anaplasmataceae agents remain unknown in wild environments in Brazil. Therefore, future studies aiming at isolating tick-borne bacteria that circulates among wildlife are much needed in order to achieve a deeply molecular and antigenic characterization of new Anaplasmataceae strains in Brazil.
ACKNOWLEDGEMENTS
We are thankful to Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) for the financial support to M.R. André (Process numbers #2015/14896-1 and #473575/2014-0, respectively), Doctorate Scholarship to K. C. M Sousa (Process #2013/13186-5)and the Scientific Initiation Fellowship to A. C. Calchi (Process # 2015/22387-0).
AUTHORS’ CONTRIBUTIONS
KCMS performed the study, carried out sampling and laboratory work, and drafted the manuscript; ACC helped with molecular analyzes; HMH helped with the animals captures in Pantanal; JSD donated Webster strain of A. phagocytophilum and prepared antigen slides; DMBB helped with arthropods identification; RZM donated Jaboticabal strain of E. canis and prepared antigen slides; MRA conceived the study and reviewed the manuscript critically for important scientific and intellectual content. All authors read, reviewed, and approved the submitted version.
DECLARATION OF INTEREST
None.
SUPPLEMENTARY MATERIAL
The supplementary material for this article can be found at https://doi.org/10.1017/S095026881700245X










