Introduction
The genus Raphanus L. [Brassicaceae, lineage 2, tribe Brassiceae (Franzke et al., Reference Franzke and Lysak2011)] is characterised, like all other members of the tribe, by heteroarthrocarpous siliques, transversally divided into two differently structured segments: a proximal, dehiscent (valvar) and a distal, indehiscent (stylar) segment (Avino et al., Reference Avino and Kramer2012). Unlike other members of the tribe, the heteroarthrocarpous silique of Raphanus is modified into a completely indehiscent, unilocular, cylindrical, thick-walled fruit, a result of the reduction of the valvar segment, and the fully developed, multi-seeded stylar segment (Schulz, Reference Schulz1919; Pistrick, Reference Pistrick1987).
In a taxonomic synopsis of Raphanus, Schulz (Reference Schulz1919) recognised two sections:
1 Section Raphanis DC., which is characterised by erect sepals, relatively long, clawed petals (1.6–2.6 cm), erect-spreading siliques and round to ovoid seeds. This section includes the polymorphic and subcosmopolitan wild radish, Raphanus raphanistrum L., under which at least two subspecies can be counted (Warwick et al., Reference Warwick and Francis2006b), the highly variable cultivated radish, R. sativus L., and the Levant-endemic R. pugioniformis Boiss., often classified as a variety of R. raphanistrum (Warwick et al., Reference Warwick and Francis2006b).
2 Section Hesperidopsis Boiss., which is characterised by patent sepals, short petals (1 cm), pendulous siliques and oblong seeds. This section is monotypic, consisting of Raphanus confusus (Greuter & Burdet) Al-Shehbaz & Warwick, formerly known by the illegitimate name R. aucheri O.E.Schulz (Al-Shehbaz & Warwick, Reference Al-Shehbaz and Warwick1997 and references therein) which is geographically restricted to northern Israel, southern Syria and Lebanon (Baillargeon, Reference Baillargeon1985a).
For the past three decades there has been much controversy over the systematic position of section Hesperidopsis. Greuter and Burdet (in Greuter & Raus, Reference Greuter and Raus1983) considered it to be a separate genus and created the binomen Quidproquo confusum Greuter & Burdet for Raphanus aucheri. Pistrick (Reference Pistrick1987), in a review of Raphanus, recognised Greuter and Burdet’s approach, and also (Baillargeon, Reference Baillargeon1985a) supported the separation of genera, mentioning that the reduced and dysfunctional valvar segment in both genera may be a result of convergence or parallelism. By contrast, Al-Shehbaz & Warwick (Reference Al-Shehbaz and Warwick1997) did not recognise the separate status of Quidproquo, arguing that differences between the proposed genera were too small to justify it. Moreover, they argued that Greuter and Burdet in Greuter & Raus (Reference Greuter and Raus1983) had not described the differences between the putative genus Quidproquo and Raphanus at generic level. Here we refer to the species at its current systematic position, namely Raphanus confusus.
To understand the taxonomic position of Raphanus confusus in relation to other Raphanus species we created a synoptic matrix of morphological characters in an attempt to evaluate similarities and differences among target taxa. We further created a phylogenetic tree based on DNA sequences of the nuclear ribosomal 5.8S and its two internal transcribed spacer (ITS) regions, and compared the molecular phylogeny with morphological data. The ITS region is the most widely used DNA locus in crucifer systematics and taxonomy (Al-Shehbaz et al., Reference Al-Shehbaz and Beilstein2006).
Method
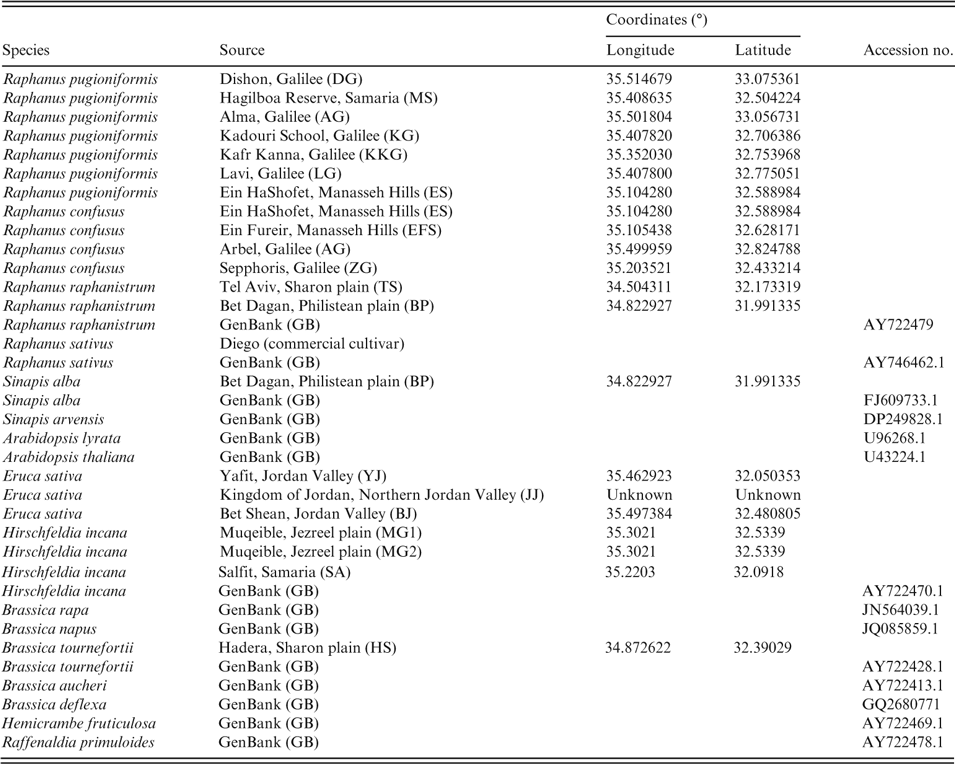
Morphological evaluation of Raphanus raphanistrum, R. pugioniformis, R. sativus and R. confusus included various morphological traits traditionally used as taxonomic markers in this group, namely fruit, flower and seed morphology plus indumentum features. For the molecular phylogenetic study, we used four recognised Raphanus species, plus reference species from several Mediterranean clades of tribe Brassiceae (Warwick & Sauder, Reference Warwick and Sauder2005); Arabidopsis (lineage 1, tribe Camelineae; Franzke et al., Reference Franzke and Lysak2011) was used as an outgroup (Table 1).
Table 1. Species and sequence data used for the current study. Accessions were either sequenced or sequences were downloaded from GenBank. Coordinates are given for accessions collected in Israel; accession numbers are given for sequences derived from the database

Leaf samples of Raphanus species were collected in their natural habitats in Israel, or species sequence information was gathered from an available database (http://www.ncbi.nlm.nih.gov/) (Table 1). Upon collection, leaf samples were placed in bags with silica gel and used for DNA isolation using an Invisorb Spin Plant Mini Kit (Stratec Molecular) following the manufacturer’s procedure.
Nucleotide variation of the nuclear 5.8S rDNA and the ITS regions 1 and 2 was used to investigate phylogenetic relationships among the Raphanus species. PCR amplifications were performed with primer sets that were designed on the basis of known sequences of the 5.8S rDNA (CGACCGACGATATGACGAG for the forward, and the degenerate GAGAACGYTSRAWCATCACTCTC for the reverse, for which: Y- C or T; S- G or C; R- A or G; W- A or T). Clear amplification products were purified using the Qiaquick PCR purification kit (Qiagene) and sequenced at the Center of Genomic Technologies (The Hebrew University of Jerusalem, Israel). Others were ligated using the CloneJET PCR (Thermo Scientific), transformed to HIT competent cells (TM-JM109, RBC) and sequenced after plasmid purification (GeneJET plasmid miniprep, Thermo Scientific).
Alignment of the 5.8S and ITS sequences was performed by the MAFFT version 7 program using default parameters (http://mafft.cbrc.jp/alignment/server/). The phylogenetic tree was then reconstructed based on the maximum-likelihood (ML) framework using the PhyML software 3.0 (http://www.hiv.lanl.gov/content/sequence/PHYML/interface.html) and the GTR model (Guindon et al., Reference Guindon and Dufayard2010). The bootstrap consensus tree (Table 2) was inferred from 100 replicates (Felsenstein, Reference Felsenstein1985), and the percentage of replicate trees in which the associated clade clustered together in the bootstrap test was designated next to the branches (Felsenstein, Reference Felsenstein1985). The tree was graphically designed with the use of FigTree version 1.4 (http://tree.bio.ed.ac.uk/software/figtree/).
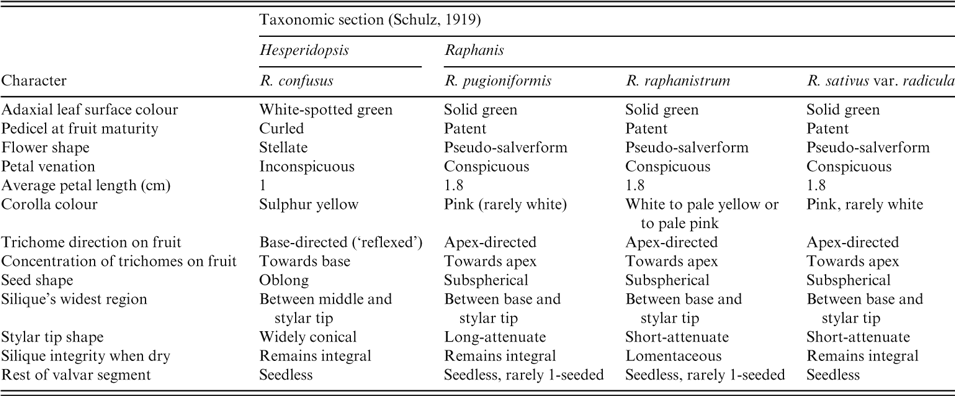
Table 2. Matrix of morphological traits in four Raphanus species according to Schulz (Reference Schulz1919) and Post (Reference Post1932)

Results and Discussion
In a matrix comparing 13 morphological traits in Raphanus species (Table 2), nine were shared by Raphanus raphanistrum, R. pugioniformis and R. sativus, but not by R. confusus. Raphanus pugioniformis, R. raphanistrum and R. sativus can be distinguished from R. confusus by their erect sepals, forming a tube-like calyx, markedly veined petals (Fig. 1), patent, attenuate fruit (thick in the middle, gradually becoming narrower), with trichomes concentrated and directed towards the apex of the fruit, and nearly spherical seeds (Fig. 2). By contrast, Raphanus confusus is distinguishable by patent sepals, unveined petals (Fig. 1), curled pedicels, pendulous siliques thickest towards the tip, silique trichomes concentrated at the base of the silique and directed towards the pedicel, and elliptical seeds (Fig. 2 and Table 2). An additional interesting unique feature in Raphanus confusus is the white spots on the adaxial leaf surface (Fig. 1).

Fig.1. Raphanus species. (A) R. confusus (= Quidproquo confusum) habit, showing white-spotted leaves; (B) R. confusus flower with patent sepals and solid-yellow petals; (C) R. pugioniformis inflorescence, showing erect sepals and dark-veined petals; (D) R. raphanistrum flower, showing dark-veined petals.

Fig.2. Fruits of Raphanus species (A) and illustration of fruit of R. confusus, indicating the direction of trichomes on the fruit (B). (See online for a colour version of this figure.)
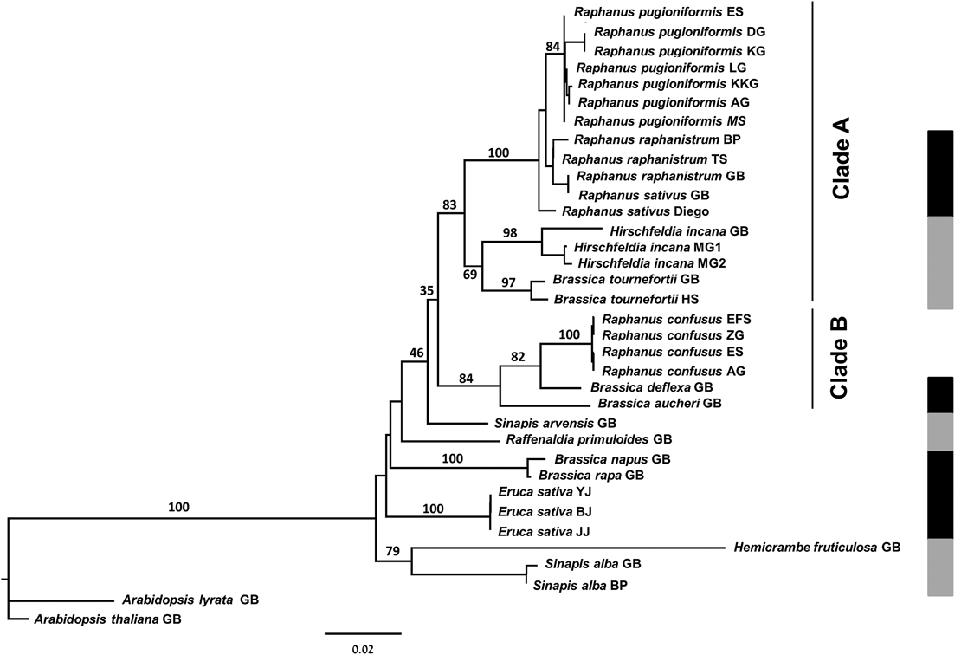
The numerous distinct characteristics of Raphanus confusus support Greuter and Burdet in Greuter & Raus (Reference Greuter and Raus1983) in questioning its systematic placement in Raphanus. Further support is provided by the phylogenetic tree based on ITS analysis which demonstrates the polyphyly of Raphanus sensu Schulz (Reference Schulz1919) (Fig. 3). The analysis placed Raphanus pugioniformis, R. raphanistrum and R. sativus (section Raphanis) as a highly supported monophyletic group (bootstrap 100) which is sister to Hirschfeldia incana and Brassica tournefortii (clade A, bootstrap 83); the monotypic section Hesperidopsis (R. confusus) is found in a highly supported monophyletic group with Brassica deflexa and B. aucheri in another lineage (clade B, bootstrap 84), which is sister to clade A (Fig. 3). The presumed close relationship between Brassica aucheri and Raphanus confusus is supported by the identical direction and location of trichomes on the silique of the two species, as can be observed in Fig. 2 and herbarium material [isotype: Iran, Haussknecht s.n., Apr. 1868 (JE!)]. This characteristic cannot be observed in Brassica deflexa, as its fruit is glabrous. A further morphological marker for clade B is the pendulous silique, which is a result of a curl (in Raphanus confusus and Brassica aucheri) or bend (in B. deflexa) of the pedicel. Moreover, in clade B, species have a strong resemblance in their flower form (stellate, due to patent sepals and petal) and colour (yellow).

Fig.3. PhyML maximum-likelihood tree of the relationships among Raphanus species. The tree is based on sequences of the nuclear ribosomal 5.8S gene and ITS region (ITS1 and partial ITS2) of the Raphanus species used in this study. Sequences of other members of the Brassicaceae were taken from GenBank (see Table 1). The bootstrap support values from 100 replicates are indicated above the branches. Arabidopsis thaliana and A. lyrata served as the outgroup. Clade A refers to Raphanus sect. Raphanis plus Hirschfeldia incana and Brassica tournefortii, and clade B to Raphanus sect. Hesperidopsis plus Brassica aucheri and B. deflexa. The panel on the right assigns the taxa studied to tribe Brassiceae lineages based on cpDNA analysis [adapted from Warwick & Black (Reference Warwick and Black1997) and Arias & Pires (Reference Arias and Pires2012)]. Black: Oleracea lineage; grey: Nigra lineage; no other Raphanus species were treated by these studies.
Our ITS results deviate from the chloroplast-based phylogenies proposed by Warwick & Hall (Reference Warwick, Hall and Gupta2009) and Arias & Pires (Reference Arias and Pires2012) and clearly show the need for more elaborate studies elucidating the phylogenetic position of Raphanus confusus. Both chloroplast-based phylogenetic trees relate Raphanus raphanistrum to the monophyletic Oleracea clade, which also includes Eruca spp., Brassica deflexa, B. napus and B. rapa, while Hirschfeldia incana and Brassica tournefortii, Raffenaldia primuloides and Hemicrambe fruticulosa are assigned to the monophyletic Nigra clade (Warwick & Hall, Reference Warwick, Hall and Gupta2009; Arias & Pires, Reference Arias and Pires2012). Conversely, both Warwick & Sauder (Reference Warwick and Sauder2005) and this present study, which are ITS-based, demonstrate that the Nigra and Oleracea lineages are non-monophyletic. Especially noteworthy is the uncertain relationship between Brassica tournefortii, Hirschfeldia incana and Raphanus raphanistrum, which appear to be closely related in the ITS analyses (Fig. 3; Warwick & Sauder, Reference Warwick and Sauder2005), but not so in cpDNA studies (Warwick & Hall, Reference Warwick, Hall and Gupta2009; Arias & Pires, Reference Arias and Pires2012). Furthermore, the relationship between Raphanus raphanistrum and Eruca spp., which is close in the cpDNA but distant in ITS, is still to be elucidated.
In the past two decades traditional taxonomic classification of the Brassicaceae has been challenged by molecular phylogenies, suggesting that many fruit characters are homoplasic (e.g. Mummenhoff et al., Reference Mummenhoff and Al-Shehbaz2005; Hall et al., Reference Hall and Tisdale2011). Furthermore, several genera which were circumscribed by a common fruit structure have been subjected to taxonomic rearrangement after molecular phylogenies had indicated a polyphyletic origin, for example Sisymbrium (Warwick et al., Reference Warwick and Al-Shehbaz2006a) and Dentaria (Sweeney & Price, Reference Sweeney and Price2000) [for a review see Al-Shehbaz (Reference Al-Shehbaz2012)].
Based on our preliminary molecular analysis, we suggest that the circumscription of Raphanus sensu Schulz (Reference Schulz1919) as well as Warwick et al. (Reference Warwick and Francis2006b) be revised. Both our ITS and the morphological data confirm that Raphanus raphanistrum, R. pugioniformis and R. sativus belong to a well-supported lineage circumscribed as Raphanus, following the generitype R. sativus (Jarvis, Reference Jarvis2007). The monotypic section Hesperidopsis, however, does not seem to belong to the same lineage as the remaining Raphanus species, and we recommend transferring it to a different genus, namely Quidproquo, reinstating the name Quidproquo confusum. Thus, section Raphanis becomes a redundant name, as it remains the only one in the genus.
Finally, our analysis supports Baillargeon’s point of view (Baillargeon, Reference Baillargeon1985a,Reference Baillargeonb), treating the reduction of the valvar fruit segment as a result of parallel evolution in the genera Raphanus and Quidproquo. Molecular analyses based upon a broad taxon sampling and a multi-gene approach are needed to assign and evaluate phylogenetic relationships of Raphanus species and related taxa of tribe Brassiceae.
Acknowledgements
This work was partially funded by the Israel Taxonomy Initiative (ITI). We thank Dr Adi Faigenboim (Agricultural Research Organization, Israel) for her assistance in the phylogenetic analysis. Thank are also due to Dr Jerry Haas and Dr Ihsan Al-Shehbaz for their input.