Ever since its discovery as a cure for pernicious anaemia, plasma levels of Cbl have been consistently identified as a modifiable endophenotype associated with an increased risk of a number of common, multifactorial diseases, including CVD, neurodegenerative disorders and different forms of cancer(Reference Green, Miller, Zempleni, Rucker, McCormick and Suttie1).
Population levels of cobalamin (Cbl) vary greatly within and between populations. This variability is explained in part by the differences in dietary intake, but also through inter-individual variability in molecular mechanisms responsible for the absorption, distribution, metabolism and elimination of Cbl. In turn, each of these processes is underpinned by the interplay of a myriad of genetic and environmental factors that remain largely unknown. By far the best studied of the molecular processes affecting Cbl disposition is Cbl metabolism, which has led to the identification of a number of genetic causes of altered Cbl metabolism including rare, severe disorders, such as inborn errors of intracellular Cbl metabolism(Reference Rosenblatt, Fenton, Scriver, Beaudet, Sly and Valle2). In addition, a more common, subtle variation in Cbl levels in the population can arise as a result of intragenic polymorphisms affecting Cbl-associated pathway(s) or intergenic regions with few obviously functional links to normal Cbl disposition, as identified recently through genome-wide association studies(Reference Hazra, Kraft and Selhub3, Reference Hazra, Kraft and Lazarus4). What has become increasingly apparent is that Cbl variability in the general population has a complex molecular underpinning, which remains largely uncharacterised.
In 1931, Garrod(Reference Garrod5) hypothesised that rare inborn errors of metabolism were merely extreme examples of what is commonly observed as more minor variations in chemical behaviour, which, in contemporary terms, would be described as conferring protection or susceptibility to diseases. So far, eight specific defects of the Cbl metabolic pathway have been identified (cblA–G and mut) and all but one (cblH) of the causative genes have been identified(Reference Coelho, Suormala and Stucki6, Reference Froese and Gravel7).
In line with Garrod's prediction, we hypothesise that common, less penetrant genetic variants at loci/genes that underlie inborn errors of Cbl metabolism are also associated with the large population variability in plasma levels of Cbl. In order to test this hypothesis and improve our understanding of the main factors responsible for population variation in Cbl levels, we carried out a classical twin study with two specific aims: (1) to characterise and quantify the genetic and environmental basis for inter-individual variation in constitutive plasma levels of Cbl and (2) to test Garrod's prediction by examining whether common genetic variations (SNP) in genes that are known to cause inborn errors of Cbl metabolism, are also candidate genes for non-pathological variation in plasma Cbl levels.
Materials and methods
Subjects
Plasma Cbl was determined in a total of 2424 twins from the TwinsUK registry (Table 1) (Reference Moayyeri, Hammond and Valdes8). All participants were of North European descent (Caucasians) and between 18 and 79 years old. We excluded twins from the study who were on any medication(s) affecting Cbl status, with a history of chronic CVD, hypertension or diabetes, or on any dietary supplements in the 3 months before participation. Prescription and over-the-counter drug and supplement use was recorded at an interview by a trained nurse. Weight and height were recorded using scales (to the nearest gram) and a stadiometer (in cm) with the subject wearing no shoes and light clothing.
Table 1 Characteristics of the study population (Mean values and standard deviations for a total of 2110 female twins)

MZ, monozygotic twins; DZ, dizygotic twins.
The present study was conducted according to the guidelines laid down in the Declaration of Helsinki, and all procedures involving human subjects/patients were approved by the St Thomas' Hospital Research Ethics Committee (EC04/015 TwinsUK). Written informed consent was obtained from all subjects.
Biochemical assays
Fasting blood samples were collected from each participant and stored on ice. Whole blood was fractionated into serum and plasma within 2 h of collection using standard procedures. Plasma samples were stored at − 20°C until assayed. Plasma Cbl and folate levels were determined by a competitive immunoassay using a direct, chemiluminescent technology(Reference Beggs, Sohn and Herrmann9). Total homocysteine (tHcy) levels were measured using isotope dilution liquid chromatography–tandem MS(Reference Ubbink, Hayward Vermaak and Bissbort10).
Preliminary statistical analysis
Equality of means and variance by zygosity
Plasma Cbl, folate, age and BMI were recorded as continuous variables. We formally tested the equality of means and standard deviations for identical or monozygotic (MZ) and non-identical or dizygotic (DZ) twins in order to determine whether the data conform to one of the assumptions of the classical twin model, i.e. there is no significant phenotypic mean or variance difference by zygosity status for the measure of interest. For this purpose, regression analyses and the equality of variance tests were conducted for age, BMI, folate and Cbl levels by zygosity to test for potentially significant differences. Observed differences by zygosity were controlled for by a quantile-normal transformation before analysis (see the ‘Genetic model fitting’ section). All preliminary analyses were performed using STATA (Stata Corporation)(11).
Covariates of cobalamin
Robust regression analyses (robust to outliers and heteroscedasticity) were used to test the effects of age, BMI, exercise, alcohol consumption, smoking status, social class and folate on Cbl levels using STATA's ‘regress’ and ‘rreg’ regression procedures. Twin-pair relatedness was accounted for in these analyses using the ‘cluster’ option in STATA.
Testing for heteroscedasticity – the relationship between cobalamin variance and age
To test for changes in Cbl variance with age, we used a mixed linear model for the regression of the between-twin variance (derived as a function of the square of the twin-pair mean(Reference Barber, Cordell and MacGregor12)) v. the fixed effects of age and age-squared. The age-squared quadratic term was included along with age to test whether the relationship was curvilinear or linear. Additional random effects were fitted for between-family variability (i.e. the intercept term specifies the family identifier) and the age slope term to estimate the population variation in both these estimates. A negative correlation between the random-effect intercept and age slope terms was modelled using STATA's ‘unstructured’ option.
Genetic model fitting
The classical twin model can be used to partition the phenotypic variance and thereby quantify the relative influence of genetic and environmental factors upon the individual variation in a population mean. The model assumes no epistasis, gene–environment correlation or interactions, and these shared environmental factors are not confounded by zygosity (the equal environments assumption). MZ twins generally share identical genes, while DZ twins share, on average, half their segregating genes. To infer heritability, therefore, greater similarity in MZ twin pairs compared with DZ pairs is attributed to genetic factors.
For estimating heritability, two standard methods are used: variance components analysis implemented using maximum likelihood(Reference Neale and Cardon13) and regression-based methods that employ the regression of the twin v. co-twin phenotype to infer variance components(Reference DeFries and Fulker14). Each method has its own advantages; thus, we used both analytical approaches in the present study. To implement a maximum-likelihood heritability model, we used the computer package Mx (www.vcu.edu/mx/), designed for the analysis of twin and family data, to partition the observed total Cbl variance (V P) into additive polygenic (V A) and dominant (V D) genetic variance component estimates and environmental components shared by both twins (V C) and unique to each twin (V E). For the analysis, we used quantile-normal-transformed Cbl values stratified by zygosity to address the sensitivity of variance components analysis to deviations from a phenotypic normal distribution(Reference Neale and Cardon13). Although regression-based DeFries & Fulker(Reference DeFries and Fulker14) heritability analyses are robust to phenotypic deviations from normality, we used the same quantile-normal-transformed Cbl phenotype both for consistency and to control for potential differences observed by zygosity. We also repeated the analyses, taking Cbl residuals for BMI before the quantile-normal transformation to check for potential confounding by zygosity.
Candidate gene study
Genotype data
Genetic data for the twins were used for the candidate gene study based on the genotypes that were already measured using the Illumina Human317-Quad for the one-half of the TwinsUK sample and the Illumina Human610-Quad Bead Chip for the other half. Illumina SNP for each candidate gene were identified for the analysis by including all intragenic SNP plus ± 20 kb (NCBI build 36) from the start and stop codons of each gene to represent core and distal regulatory regions.
Quality control
For quality control, two levels were used(Reference Weale15). First, subjects were excluded based on the genotype data when (1) the SNP call rate was < 95 % (missingness) and (2) heterozygosity was >37 or < 33 % across all SNP, which is a measure of the expected genetic variation in a population in the absence of any systematic genotyping or sample-handling errors. The samples were tested for population stratification using multidimensional scaling, by comparison with three HapMap phase 2 reference populations (CEU (Utah residents with Northern and Western European ancestry), YRI (Yoruban) and CHB/JPT (Han Chinese in Beijing, China/Japanese)). Ethnic outliers were identified and excluded. Second, SNP were removed if they deviated from Hardy–Weinberg equilibrium at P≤ 1 × 10− 4, a minor allele frequency ≤ 1 % and a SNP call rate ≤ 95 %.
Selection of SNP in candidate gene regions
We used the ‘tagger’ tool in Haploview software(Reference Barrett, Fry and Maller16) to quantitatively assess how well the available genotype data represented, i.e. tagged, all known common genetic variations in each gene using a pairwise r 2 metric threshold ≥ 0·8 (referred to as tagging efficiency). The degree of linkage disequilibrium –or the extent to which small genomic regions historically co-segregate (and are associated) with one another – in each gene region was also assessed by plotting the pairwise correlation for all the genotyped SNP for each gene (data not shown).
Genetic association analysis
Tests of the genetic association between Cbl levels and each SNP were conducted using GenABEL implemented within the statistical package R(Reference Aulchenko, Ripke and Isaacs17). Briefly, each SNP was tested for the association with Cbl levels using an additive genetic model(Reference Falconer and Mackay18) in which each SNP is coded 0, 1 or 2, reflecting a count of the number of copies of the minor allele observed at each diploid locus. For each test, we used a likelihood ratio test to compare a model that included a SNP with a null model in which the SNP is removed from the regression. The likelihood ratio test provides a statistical χ2 test of significance with one df for the additive genetic model. Relatedness among twins was accounted for using a weighted analysis based on a pairwise kinship coefficient matrix for all the study subjects calculated using the genome-wide Illumina panel of SNP.
Multiple testing
The type I error rate was controlled for using a Bonferroni correction with the adjusted statistical threshold of significance α′= α/n, where α is the nominal significance threshold for a single test (conventionally α = 0·05) and n equals the total number of genotyped SNP tested. The total number of genotyped SNP included in the present study was thirty-eight with a total of eight genes tested, which corresponds to a conservative significance threshold of α′ = 0·05/38 = 0·001.
Results
Characteristics and blood measurements of the study participants are summarised in Table 1. We excluded the data from male twins (n 150) due to inadequate power as well as twins who suffered from any chronic diseases (n 164). A total of 2110 female twins (262 MZ, 784 DZ twin pairs and nineteen unknown zygosity singletons) were included in the study. None of the participants was treated with drugs known to interfere with Cbl uptake, including anticonvulsants, peptic ulcer drugs, antimetabolites, trimethoprim, hydrazides and mestoranum, or with B-complex supplements.
For each participant, we had data for age, BMI, self-report questionnaire data on alcohol use, smoking status and employment (used to derive social class using the UK General Registrar Classification), Cbl, tHcy and folate levels. These twins represented about one-third of the females on the TwinsUK database. Although they had a mean age that is approximately 5 years older than the remaining TwinsUK database volunteers, they were comparable for mean weight, prevalence of smoking, levels of alcohol consumption as well as a number of other demographic and lifestyle variables. Furthermore, the Cbl sample had a slightly lower prevalence of smoking and alcohol consumption compared with other age-matched populations(Reference Andrew, Hart and Snieder19).
Relationship between age, BMI, exercise, alcohol consumption, smoking status, social class, folate, total homocysteine and cobalamin levels
We identified a total of thirty individuals who were putatively Cbl deficient (Cbl < 150 pmol/l), despite this group having the same mean age and age range compared with the rest of the sample (18–79 years old). Altogether, 88 % of the participants had a Cbl value within the normal range (150–660 pmol/l). As elevated levels of Cbl (>660 pmol/l) are uncommon and may be due to liver disease, we checked liver function test protein levels (alanine transaminase, γ-glutamyl transpeptidase and alkaline phosphatase) for 241 women with Cbl values above 660 pmol/l and found them to be all normal and within the normal range (data not shown).
Small differences in mean and variance for Cbl levels between MZ and DZ twin pairs were identified and controlled for in the heritability analyses by transforming Cbl values using a quantile-normal function stratified by zygosity. Age (β = 7·5) and age-squared (β = − 0·1, P= 7 × 10− 7, R 2< 0·02) and BMI (β = − 2·3, P= 3 × 10− 4, R 2< 0·006) were shown to have a small, but significant effect on Cbl levels with BMI showing a negative and age a positive relationship with Cbl levels (Table 2). Women who reported taking regular physical exercise in the previous year (11 % of all female twins), on average, had Cbl levels that were 30 pmol/l higher than those who reported no regular exercise (P= 0·02, R 2 0·004). Similarly for alcohol consumption, women who self-reported either as never or drinking less than 5 units/week (73 %) had plasma Cbl levels that were, on average, 32 pmol/l higher than those who reported drinking between 5 and 40 units/week (P= 0·02, R 2 0·005). Women who reported never having smoked (55 %) had Cbl levels that were on average 35 and 21 pmol/l higher than those who were current (18 %) and ex-smokers (28 %), respectively (P= 0·09, R 2 0·005).
Table 2 Association of plasma cobalamin (Cbl) levels with age, BMI, self-reported current physical activity, current alcohol consumption, smoking status, social class and plasma folate levels

β, Regression coefficient; se, robust standard error; R 2, regression coefficient of determination (i.e. proportion of the Cbl variance explained by the model).
* The model of Cbl regression with age includes a quadratic age term (age2) to account for the curvilinear relationship between Cbl levels and age in the population.
† Physical activity is recorded as self-reported (yes/no) regular activity in the previous year.
‡ Alcohol consumption is modelled as ‘never’ or ‘1–5 units/week’ v. ‘>5 (5–40) units/week’.
§ Smoking status is modelled as ‘never smoked’ v. ‘current’ and ‘ex-smoker’ categories.
∥ Social class is based on the UK General Registrar Classification with the categories of senior management (A) or professional (B), skilled non-manual (C1) or skilled manual (C2) v. semi- (D) or unskilled worker (E).
Based on their employment status, women who were classified (UK General Registrar Classification) as being social class D/E (semi-skilled or unskilled) had Cbl levels that were on average 48 and 72 pmol/l lower than the social class groups A/B (senior management/professional) and C1/C2 (skilled non-manual/skilled manual, P= 0·02, R 2< 0·01), respectively. Folate levels were strongly and positively associated with the variation in Cbl levels (β = 8·9, P= 8 × 10− 4, R 2< 0·14; Table 2). Multiple regression analysis confirmed that age, BMI and folate remained associated with Cbl levels, collectively accounting for approximately 15 % of the variation in Cbl levels (data not shown).
Effect of age on cobalamin levels
Previous studies have shown a clear increase in Cbl deficiency and insufficiency in individuals over the age of 60 years due to the decreased absorption of protein-bound Cbl in the elderly(Reference Baik and Russell20). We sought to verify the relationship between Cbl levels and age in the present data. Scatter plots illustrated the positive curvilinear relationship between plasma Cbl levels and age and the large variation in Cbl levels for all ages in this population-based sample of female twins. Despite the large variation, average plasma Cbl levels (fitted line) were seen to increase modestly from the age of 18 years until approximately the mid-40s, plateau and then decrease from the age of 60 years (data not shown).
To assess heteroscedascity (i.e. the change in Cbl variance across the range of an explanatory variable), we initially plotted the standard deviation of Cbl against arbitrarily defined age bins, and observed a positive increase in phenotypic variance with age (data not shown). When we formally tested this relationship using a mixed model to regress between-twin Cbl variance upon age and age-squared, we confirmed a significant (χ22= 12, P= 0·003) curvilinear relationship with the variance in Cbl, declining from 18 years to mid-40s and increasing again after the age of 50 years.
Genetic model fitting and estimation of heritability
The results of genetic model fitting for the quantile-normalised Cbl data are shown in Table 3. The most parsimonious and best-fitting model included an additive polygenic and unique environmental variance component (AE model), with up to 53 % of the variation in plasma Cbl levels accounted for by additive genetic factors (i.e. heritability). The remainder of the variance was accounted for by environmental factors unique (non-shared) to each twin. Given the relative lack of power to detect shared familial effects in twin studies per se (Reference Hopper21), however, the present data also provide evidence that up to 10 % of the variance in plasma Cbl levels is attributable to shared familial environmental factors, when an ACE model is fitted, with the ACE model including additive polygenic (A), common (C) and unique (E) environmental effects.
Table 3 Heritability of plasma cobalamin levels

Δχ2, change in χ2 fit statistic or likelihood ratio test (LRT); A, additive polygenic variance component; C, shared familial or common environment; D, dominant genetic variance; E, unique environmental effects (including measurement error) that are specific to the individual.
* Five models are presented, each with best-fit model statistics and standardised variance component estimates for A, C, D and E that add up to one for each model. The most parsimonious and best-fit model (AE) is highlighted, with a heritability estimate of 0·53 (95 % CI 0·47, 0·59) and the remaining variance (0·47) attributed to environmental factors specific to the individual. Model-fit comparisons are based on a LRT, where the model-fit contribution for each parameter (A, D and C) is assessed by individually fixing the value of each to zero to contrast the full model (ACE or ADE) with a nested sub-model (AE, CE and E). If the LRT is significant (here assessed by the change in χ2 with one df), the variance component contributes to the model fit and cannot be dropped. Common environment (C) and dominant genetic (D) effects are confounded using twin heritability models, so these components cannot be estimated in the same model.
Given the observation that the mean and variance for the Cbl increase with age, we also fitted heritability models stratified by three age groups (under 40, 40–55 and over 55 years). The Defries–Fulker regression-based models suggest that the genetic variance of Cbl increases with age with heritability for each group being 18 % (ACE model), 48 % (AE model) and 58 % (AE model), for the under 40-, 40–55- and over 55-year age groups, respectively (Table 4). It should be noted that only the under 40-year age group provided good evidence for a model that includes a large shared familial component (ACE) estimated to account for 30 % of the total phenotypic variation in plasma Cbl for this age group. The middle-aged and elderly group best-fit models did not provide evidence for shared familial components (with the best-fit model for both these age groups being the AE models).
Table 4 Change in the heritability of plasma cobalamin levels by age

n, Number of participants; A, additive polygenic effects; C, shared familial or common environmental effects; D, dominant genetic variance; E, unique environmental effects (including measurement error) that are specific to the individual; ACE, model including variance component (VC) estimates for additive polygenic, shared familial and unique environmental effects; CE, model including only VC for shared familial and unique environmental effects; AE, model including additive polygenic and unique environmental effects; adj R 2, adjusted R 2 model best-fit statistic.
* Three VC models are presented for each age group, each with best-fit statistics and VC estimates that add up to one. The best-fit model (based on both the adjusted R 2 fit statistic and the model providing interpretable, non-negative VC estimates) is ACE for the under 40-year age group and AE for the middle-aged and older age groups. Heritability estimates are based on DeFries–Fulker regression analyses of twin data(14), in which unlike VC analyses, regression-based VC estimates are not constrained to lie between 0 and 1, but are freely estimated.
Impact of inborn errors of cobalamin metabolism genes on constitutive plasma cobalamin levels
Tagging efficiency
We formally quantified how well HapMap common SNP, with a minor allele frequency ≥ 10 %, were captured and represented by our sample genotyped SNP based on the degree of correlation (r 2) between each SNP pair (namely tagged and tagging SNP, respectively). Based on a minimum r 2 of 0·8, Table 5 shows that our genotyped SNP provided a reasonable representation for most of the genes with the percentage of common SNP tagged ranging from 40 to 92 %. For gene regions MMACHC and MMAA (methylmalonic aciduria cbIC type, with homocystinuria and methylmalonic aciduria (cobalamin deficient) cbIA type), the proportion of SNP not represented was high with half or more of the common variants not adequately captured. Excluding these two gene regions, the mean proportion of common variants captured by our panel of genotyped markers was close to 80 %.
Table 5 SNP marker coverage for each of the eight candidate genes

cbl, Cobalamin.
* The number of genotyped SNP for each gene region.
† The degree to which these markers are pairwise correlated (r 2) with all common SNP documented by the HapMap project for these regions.
Genetic association analysis
We tested whether common SNP in genes known to cause Mendelian defects in inborn errors of Cbl metabolism and transport are associated with plasma Cbl levels. A total of thirty-eight genotyped SNP– tagging a total of 255 common (minor allele frequency ≥ 0·10) known SNP (minimum r 2≥ 0·8) documented by HapMap – were identified in eight gene regions and tested for the association with plasma levels of Cbl in female Caucasian twins from the TwinsUK registry. It should be noted that for these analyses, we used only one individual from each MZ twin pair with the most complete phenotype data.
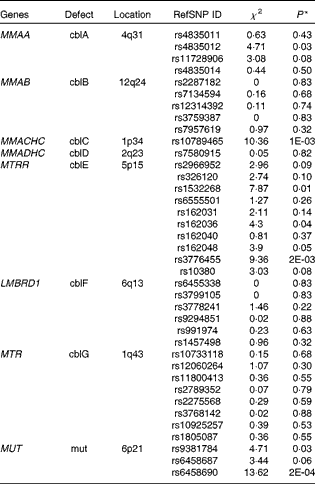
The results of the analyses are shown in Table 6. Common polymorphisms in the genes MMAA, MMACHC, MTRR (5-methyltetrahydrofolate-homocysteine methyltransferase) and MUT (methylmalonyl coenzyme A mutase) were observed to be nominally associated (α = 0·05) with plasma Cbl levels. MMACHC (P= 0·001) and MUT (P= 0·0002) both contained SNP associated with Cbl that passed the conservative Bonferroni significance threshold adjusted for multiple testing (α′ = 0·001). Furthermore, multiple variants in two of the four genes, MTRR and MUT, showed evidence for the association with Cbl levels, strengthening the case for the importance of these genes in potentially driving population variability in Cbl levels. We failed to show the association between common variants in the genes MMAB, MMADHC, LMBRD1 and MTR (methylmalonic aciduria (cobalamin deficiency) cbIB type, methylmalonic aciduria cbID type, with homocystinuria, LMBR1 domain containing 1 and methyltransferase reductase) and plasma levels of Cbl.
Table 6 Results from the candidate gene association study

RefSNP ID, reference SNP ID number; χ2, chi-squared statistic; cbl, cobalamin.
* Significant association with Cbl levels was based on a conservative Bonferroni corrected P≤ 0·001.
Discussion
We aimed to gain a better understanding of the main genetic and environmental factors that affect the population variation in plasma Cbl levels using a classical twin model. We also tested Garrod's hypothesis that common, less penetrant genetic polymorphisms in genes that underlie inborn errors of Cbl metabolism and transport may be associated with normal population variability in plasma Cbl levels.
The present results verify that variations in plasma levels of Cbl are highly heritable. To our knowledge, this is the first study to report heritability for Cbl levels (estimated to be approximately 53 %) for a female population. The only other report is from a much smaller study of ninety-six MZ and 120 DZ twin pairs conducted in elderly Swedish twins (age ≥ 82 years), which reported a heritability of 62 % for serum Cbl levels(Reference Nilsson, Read and Berg22).
The difference between the heritability estimates between the present study and the Swedish study is likely to be attributable to the large, non-overlapping difference in age for the two samples, with all study subjects under 80 years of age for the TwinsUK and over 80 years of age for the Swedish study. The reported mean Cbl values for the two studies are also appreciably different with the TwinsUK value (overall female mean 422 pmol/l) up to 20 % higher than those reported by the Swedish study (302 and 344 pmol/l for men and women, respectively). It is worth highlighting that the (1) difference in mean plasma Cbl levels between the two studies could be in part due to differences in the assays used to measure Cbl levels in each study, and (2) heritability estimates between the two studies diminished considerably when comparing the Swedish study with the results from the TwinsUK participants aged over 55 years. For this age group, the heritability estimate is 58 %, which is comparable with the Swedish Cbl heritability estimate of 62 %.
The present findings support data from the literature showing that plasma cbl levels are significantly correlated with age, BMI, alcohol use, smoking status, social class and folate levels, which in our cohort collectively explained up to 15 % of the variation in Cbl levels. Importantly, the present results lend further support to the observational link between the increasing Cbl deficiencies in the elderly, which is commonly attributed to the increased prevalence of atrophic gastritis in this age group and shows both the prominence of genetic factors in driving increased variation in Cbl levels with age and the importance of shared environmental factors – for example, similar dietary habits in earlier life – for younger age groups. This highlights the need to better manage Cbl deficiencies in the elderly through an intervention programme that more specifically targets this stratum of the population.
Finally, the results from the present candidate gene study show the potential for studying genes that underlie rare, Mendelian diseases of Cbl metabolism and transport in the context of normal population variability in plasma Cbl levels. Out of the eight genes investigated, four – including MMAA, MMACHC, MTRR and MUT – harbour common polymorphisms that were either suggestive or significantly associated with Cbl levels. This underlines the continuing importance of formulating study designs to test hypotheses where a strong body of evidence already exists – for example, Mendelian conditions with pathophysiological markers – since the observed associations for MMAA, MMACHC and MTRR common polymorphisms in the present study would have probably been overlooked by Genome-wide Association Study (GWAS) studies. In the case of MUT, the strongest association in the present study is with SNP rs6458690 (P= 0·0002), which is in perfect linkage disequilibrium with two SNP (r 2 1), rs9473558 and rs9473555. Both of these SNP in MUT have previously been shown to be strongly associated with plasma levels of Cbl(Reference Hazra, Kraft and Lazarus4) and tHcy levels(Reference Paré, Chasman and Parker23). Moreover, power constraints may have contributed to our failure to show the association between MTR and Cbl levels, as SNP rs2275565 in MTR is observed to be strongly associated with tHcy levels in larger studies (JBJ van Meurs, G Pare, SM Schwartz, et al. unpublished results). Although this SNP was not included in the genotype panel, it is estimated to be in strong pairwise linkage disequilibrium (r 2 0·86, HapMap CEU data) with two MTR SNP that are genotyped for the present study, rs10925257 and rs1805087.
The present results lend support to Garrod's prediction that rare, inborn errors of metabolism are extreme examples of what is commonly observed as minor, healthy variations in chemical behaviour. Inborn errors represent a large number of rare metabolic diseases that affect every major organ system with diverse manifestations. It has been estimated that inborn errors of metabolism can account for up to 15 % of all single gene disorders in the population, representing a significant number of genes to be interrogated with suitable biochemical phenotypes, as illustrated in the present study. Once identified, this approach might facilitate the remedy for micronutrient imbalances through, for example, appropriate supplementation of the cognate marker(Reference Ames, Elson-Schwab and Silver24).
Potential limitations
The present study has a few potential limitations, which warrant comment. First, although we aimed to include twins in the study who were deemed to be largely healthy (assessment through general health questionnaires, multiple clinical visits, not taking prescribed medication for chronic diseases), the present results may have nevertheless been influenced to an unknown extent by undocumented circumstances such as irregular diet, mild infection, early stages of a disease or a short course of an over-the-counter medication.
Another limitation was the sex constituency of the study participants. Due to historical reasons, TwinsUK is predominantly made up of female twin volunteers. Although we had data on Cbl levels for up to 200 male twins, we felt that the small sample size and the unbalanced design severely limited our ability to reliably comment either on men or sex differences, so as a result, we did not include the available males with Cbl measures in the present study. This is important in light of well-documented sex differences in constitutive tHcy levels, a marker that is directly influenced by Cbl levels.
The present results are also constrained by the power of the study. The present study had high power to detect low heritability and to test and quantify the effects of potential covariates consistent with previously published results in the literature. Nevertheless, the present genetic association study was underpowered for some of the candidate genes with poor marker coverage (e.g. for regions of low linkage disequilibrium including MMAA, MMACHC, MMADHC and MTRR) or the candidate genes harbouring associated variants, but with low penetrance (regardless of reasonable marker coverage). Similarly, the lack of a replication study/cohort is a limitation, which would reduce the potential for type I errors. Nevertheless, the fact that some of the present results replicate the findings from other studies adds confidence to our observations.
Finally, given the findings in the older age group, it would also be important to test the influence of the higher prevalence of impaired renal function and/or Cbl absorption on the present results. Similarly, it is important to validate the results observed for the present study using other biomarkers of Cbl status such as methylmalonic acid and holotranscobalamin.
In conclusion, the present results show that the observed population variability in Cbl levels is driven by a large number of genetic and environmental factors that contribute differentially as the population ages. Interestingly, we showed that genetic factors are responsible for the increase in plasma Cbl variance with age. This novel observation warrants further investigation, as it may be important to know what genetic factors are driving the increase in phenotypic variance as the population ages. Finally, in line with Garrod's prediction, we observed that common genetic variants in genes that underpin rare Mendelian forms of inborn errors of the Cbl metabolic pathway are also associated with and contribute to the genetic basis for the variation in Cbl levels. While the importance of Cbl metabolic pathway genes in determining Cbl deficiency in old age remains to be determined, the present study does illustrate that at least some of these genes play a role in regulating Cbl levels in the general population.
Acknowledgements
The authors would like to thank all the twins who participated in the study and acknowledge the dedication and support of the personnel at the Department of Twin Research and Genetic Epidemiology. T. A. acknowledges support by the Medical Research Council UK (Investigator Award 91993).
The authors acknowledge support from the Chronic Diseases Research Foundation, the Medical Research Council and the Wellcome Trust.
K. R. A. conceived the study. K. R. A. and T. A. designed and supervised the study. T. A. and K. R. A. conducted the research. R. G. and I. G.-N. were responsible for the recruitment and phenotypic data queries and management. T. A. analysed the data. K. R. A. and T. A. wrote the paper and had primary responsibility for the final content. All authors read and approved the final manuscript.
There are no conflicts of interest to declare.