Diet-induced milk fat depression (MFD) is caused by the inhibition of milk fat synthesis by bioactive fatty acids (FA) synthesised by rumen microbes, and is a well-studied example of the interaction between dietary nutrients, the gastrointestinal microbiome and tissue physiology. Rumen microbes perform a wide range of functions and are classically grouped within niches based on their predominant substrate or enzyme activity. Moreover, rumen microbes biohydrogenate unsaturated FA, resulting in the formation of trans isomers as intermediates. The rate, extent and pathways of biohydrogenation (BH) are commonly attributed to the microbial population present in the rumen. Previous investigations quantifying ruminal microbial populations have been conducted after diet adaptation periods; however, the time course of adaptation to a new diet is not well characterised. Importantly, investigation of the time course is essential to establish the potential for the changes in the microbial population as a primary mechanism in diet-induced milk fat depression, rather than simply as a secondary adaptation.
Milk FA are a sensitive indicator of absorbed FA profile, as 85 % of preformed FA originate directly from intestinal absorption in cows in positive energy balance( Reference Palmquist, Fox and McSweeney 1 ). Shingfield et al. ( Reference Shingfield, Reynolds and Hervas 2 ) and Rico & Harvatine( Reference Rico and Harvatine 3 ) reported the time course of the changes in milk FA profile when cows were switched from a control diet to a diet that resulted in MFD because of high diet fermentability and high PUFA concentration. During the transition period, a biphasic response in BH intermediates was observed( Reference Rico and Harvatine 3 ). The rate or capacity of the normal rumen BH pathway was first decreased, causing a 2- and 3-fold increase in trans-11 18 : 1 and cis-9, trans-11-conjugated linoleic acid (CLA) on days 3 and 5, respectively. After day 5, these intermediates decreased rapidly. At the same time, there was a progressive increase in the alternative BH pathway that became predominant after 9 d as indicated by an increase in the production of trans-10 18 : 1 and trans-10, cis-12-CLA. Similar temporal shifts in microbial populations are expected to be the driver of these metabolic changes.
Rumen microbes are sensitive to dietary risk factors for MFD including diet fermentability and unsaturated FA concentration, low ruminal pH, and monensin. For example, Weimer et al. ( Reference Weimer, Stevenson and Mertens 4 ) reported broad changes in ruminal bacterial communities when feeding high-starch and monensin-supplemented diets using automated ribosomal intergenic spacer analysis, and others have reported changes in specific taxa by real-time quantitative PCR( Reference Tajima, Aminov and Nagamine 5 – Reference Mullins, Mamedova and Carpenter 7 ). Furthermore, Rico et al. ( Reference Rico, Ying and Clarke 8 ) reported that inoculation of MFD cows with rumen digesta from cows fed a high-fibre and low-PUFA diet slightly accelerated the recovery of normal ruminal BH intermediates in MFD cows, thus suggesting that re-establishment of a normal ruminal microbial population may limit the rate of recovery from MFD.
The objective of the present study was to determine the time course of the shift in the rumen microbiome during dietary changes by investigating selected microbial taxa with well-characterised functions using quantitative PCR, which is a sensitive method to detect changes( Reference Kobayashi, Koike and Taguchi 9 ). Similarly to Mullins et al. ( Reference Mullins, Mamedova and Carpenter 7 ), we propose that the selected taxa would provide a proxy of general changes in the microbial ecosystem. The classic culturable bacterial species represent only a small fraction of the total rumen microbiome( Reference Stevenson and Weimer 6 ); however, specific well-characterised bacterial species are expected to represent the niche they occupy in the rumen and to demonstrate the temporal adaptation of the rumen. Additionally, the ruminant is an interesting model for studying the gastrointestinal microbiome, as cannulation allows direct sampling of the most active fermentation compartment. We hypothesised that rumen microbes would rapidly adapt after a dietary switch, and that the dynamics of rumen taxa would correlate with changes in the profile of milk trans-FA.
Materials and methods
Experimental design
All experimental procedures were approved by the Pennsylvania State University Institutional Animal Care and Use Committee. All cows were housed in a tie-stall barn located at the Pennsylvania State University's Dairy Production Research and Teaching Center, and fed individually once daily (08.00 hours) at 110 % of expected intake.
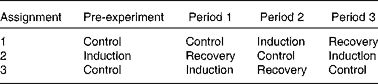
A total of eight ruminally cannulated cows were used to investigate the time course of the induction of and recovery from MFD in a replicated design with three periods of 21 d (one cow failed to reduce milk fat and was not included in the experiment). A detailed description of the experimental design and treatments has been reported previously( Reference Rico and Harvatine 3 ). Briefly, in each treatment sequence, induction of MFD followed the control period, and recovery from MFD followed the induction period (Table 1). A pre-trial period was necessary to provide an induction period before recovery in period 1 for treatment sequence 2. Induction of milk fat depression was achieved by feeding cows a low-fibre, high-PUFA diet (29·5 % neutral-detergent fibre (NDF), 27 % starch, 5·5 % FA and 3·7 % PUFA; DM basis) for a period of 21 d. Following the induction of MFD, dairy cows were switched to a high-fibre, low-PUFA diet (36·9 % NDF, 18 % starch, 2·6 % FA and 1·1 % PUFA) to observe recovery of milk fat for 21 d.
Table 1 Treatment assignment of a repeated design to study the induction of and recovery from diet-induced milk fat depression
Sampling procedures
Whole rumen digesta samples were collected at 16.00 hours on days 0, 4, 8, 12 and 20 of each experimental period. The sample was collected approximately 8 h after feeding, and represents the high intake and rapid fermentation period of the day. Digesta samples were collected from five different locations of the rumen (cranial dorsal, cranial ventral, central, caudal dorsal and caudal ventral), composited, subsampled (approximately 250 g) and stored at − 20°C. The samples were freeze-dried (Ultra 35-XL; Virtis Company, Inc.) and ground in a coffee grinder (Model 80 335; Hamilton Beach Brands, Inc.) for 60 s before DNA extraction. Importantly, the procedures used herein provided whole digesta samples representing the microbial populations in both the liquid and the solid fractions (see Mullins et al. ( Reference Mullins, Mamedova and Carpenter 7 )).
DNA extraction and quantitative PCR analysis
DNA was isolated once for each cow at each time point using a commercially available kit (QIAamp DNA Stool Mini Kit; Qiagen Sciences) with modifications similar to those proposed by Yu & Morrison( Reference Yu and Morrison 10 ). Briefly, approximately 220 mg of freeze-dried and ground digesta were homogenised for 5 min with 1·2 ml lysis buffer and 0·4 g of 0·1 mm sterile zirconia beads using a bench-top vortex equipped with a MO BIO Vortex-Genie 2 adapter (MO BIO Laboratories). A mixture of 0·5 and 0·1 mm beads, as proposed by Yu & Morrison( Reference Yu and Morrison 10 ), was tested, but did not improve the yield. After bead-beating, the samples were incubated twice at 95°C for 15 min with vortexing every 5 min and bead-beating between the incubation periods. Lastly, DNA was purified by column extraction, and DNA concentration was determined by spectroscopy (NanoDrop ND-1000 Spectrophotometer; NanoDrop Technologies). The repeated bead-beating and the column purification in the extraction method provided high yield and quality of DNA( Reference Yu and Morrison 10 , Reference Henderson, Cox and Kittelmann 11 ). The relative abundances of lactate-utilising bacteria (Megasphaera elsdenii and Selenomonas ruminantium), amylolytic bacteria (Streptococcus bovis and Prevotella bryantii), fibrolytic bacteria (Ruminococcus albus, Fibrobacter succinogenes, and Prevotella ruminicola), bacteria involved in BH (trans-11 18 : 1 producers, the Butyrivibrio fibrisolvens/Pseudobutyrivibrio group and Butyrivibrio hungatei), anaerobic fungi (Neocallimastigales) and ciliate protozoa were determined in triplicate using real-time quantitative PCR and previously validated primers and conditions (400 nm of each primer; see online Supplementary Table S1). Reactions were performed using a commercial mix (PerfeCTa SYBR Green SuperMix with ROX; Quanta Biosciences), and amplification fluorescence was measured (Applied Biosystems 7900 HT Fast Real-Time PCR System; Life Technologies). Primer specificity was evaluated by melting curve analysis, and efficiency (E) was calculated as E= 10− 1/slope of the standard curve. Efficiency values ranged between 1·86 and 2·27 (mean 2·04). Abundance of bacterial species are expressed as a percentage of total bacteria, calculated as E(UB) C t(UB)/E(TG) C t(TG), where E(UB) is the efficiency of the reaction for the universal bacterial primer set; E(TG) is the efficiency of the reaction for the target gene; and C t is the average C t value for each sample. Abundance of total fungi, protozoa and bacteria are expressed relative to day 0 of the control period (after feeding the low-fibre, high-PUFA diet for 20 d) and relative to total bacteria, which may account for the differences in DNA integrity and possible PCR inhibitors.
Statistical analysis
Data were analysed using the MIXED procedure of SAS with repeated measures (version 9.3; SAS Institute, Inc.), as described previously( Reference Rico and Harvatine 3 ). Briefly, the model included sequence and period as random effects, and treatment, time and their interaction as fixed effects. Time was used as the repeated variable and cow by treatment as the subject. The Kenward–Rogers method was used for the adjustment of the denominator df. Furthermore, first-order antedependence covariance structure (allowing unequal spacing) was used in the model. Individual pre-planned contrasts tested control v. induction and control v. recovery at each time point. Significance and tendencies of main effects and pre-planned contrasts were set at P< 0·05 and P< 0·10, respectively, and interactions at P< 0·10 and P< 0·15, respectively. Data were log-transformed (required for all microbial populations), and back-transformed least-squares means and the largest standard error of the means are reported. Second, a random regression model was used to evaluate the associations between rumen microbes and milk fat concentration and milk FA profile, between milk fat concentration and milk FA profile, and between individual groups of rumen microbes from observations on days 0, 5, 9, 13 and 21 using JMP Pro (version 10.0.0; SAS Institute, Inc.). The full model included cow and period as random effects and each microbial taxa as linear and quadratic effects. The quadratic effect was removed from the model when P>0·05 only if the model fit improved based on the Akaike information criterion. Associations with a coefficient of determination >0·40 are discussed.
Results
Milk production and milk FA profile have been reported previously( Reference Rico and Harvatine 3 ). Briefly, milk yield was not affected by treatment, but milk fat yield progressively decreased during the induction period and was 23 % lower than the control levels by 7 d and was 34 % lower from day 9 to 21. Milk fat yield progressively increased when dairy cows were fed the recovery diet and were not different from that of the control group on day 15. A biphasic response was observed for milk fat trans isomers, where the concentrations of trans-11 18 : 1 and cis-9, trans-11-CLA were elevated initially and those of trans-10 18 : 1 and trans-10, cis-12-CLA increased progressively during the induction period. Similarly, a biphasic response was observed during the recovery from MFD, with the concentrations of trans-10 18 : 1 and trans-10, cis-12-CLA rapidly decreasing initially and those of trans-11 18 : 1 and cis-9, trans-11-CLA increasing slightly above the control levels during the second phase of recovery. Treatment × time interactions were detected for all microbial taxa quantified (P< 0·05), except for P. ruminicola and B. hungatei.
Fungi, ciliate protozoa and total bacteria
Total abundance of fungi, ciliate protozoa and bacteria were first determined to observe major shifts in the concentration of rumen microbes (Fig. 1). Fungal abundance decreased by 93 % on day 4 of induction, when it reached a nadir (P< 0·01; Fig. 1(a)). Conversely, during the recovery period, the abundance of fungi increased progressively, was not different from the control levels on day 8, and reached a plateau on day 12. Similarly, the induction diet progressively decreased the relative abundance of ciliate protozoa, which tended to be lower than the control levels on day 4 (P= 0·09; Fig. 1(b)) and reached a nadir on day 8 at a 92 % reduction compared with the control diet (P< 0·01). Similar trends were observed for fungi and ciliate protozoa relative to total bacteria (see online Supplementary Fig. S1)

Fig. 1 Time course of the total abundance of (a) fungi, (b) ciliate protozoa and (c) bacteria during the induction of and recovery from milk fat depression. Data are plotted relative to day 0 of the control period. Dairy cows were fed a high-fibre, low-PUFA diet (control; ![]() ), a low-fibre, high-PUFA diet resulting in milk fat depression (induction;
), a low-fibre, high-PUFA diet resulting in milk fat depression (induction; ![]() ), or a high-fibre, low-PUFA diet following induction that resolved milk fat depression (recovery;
), or a high-fibre, low-PUFA diet following induction that resolved milk fat depression (recovery; ![]() ). Values are least-squares means, with their largest standard errors of the mean ((a) sem 0·64, (b) sem 0·91 and (c) sem 0·23). Mean value for the induction group was significantly different from that of the control group: * P< 0·10, ** P< 0·05, *** P< 0·01 (pre-planned contrasts). Mean value for the recovery group was significantly different from that of the control group: † P< 0·10, †† P< 0·05, ††† P< 0·01 (pre-planned contrasts). For fungi and ciliate protozoa, there was a significant main effect of treatment (P< 0·001 both), but no effect for time (P= 0·45 and P= 0·54, respectively), and the treatment × time effect was significant (P< 0·001 both). For bacteria, there was a significant treatment × time effect (P= 0·02), but no effects for treatment (P= 0·21) and time (P= 0·38).
). Values are least-squares means, with their largest standard errors of the mean ((a) sem 0·64, (b) sem 0·91 and (c) sem 0·23). Mean value for the induction group was significantly different from that of the control group: * P< 0·10, ** P< 0·05, *** P< 0·01 (pre-planned contrasts). Mean value for the recovery group was significantly different from that of the control group: † P< 0·10, †† P< 0·05, ††† P< 0·01 (pre-planned contrasts). For fungi and ciliate protozoa, there was a significant main effect of treatment (P< 0·001 both), but no effect for time (P= 0·45 and P= 0·54, respectively), and the treatment × time effect was significant (P< 0·001 both). For bacteria, there was a significant treatment × time effect (P= 0·02), but no effects for treatment (P= 0·21) and time (P= 0·38).
There was no overall effect of treatment on the relative abundance of total bacteria, which remained relatively constant during the induction and recovery periods (Fig. 1(c)). However, during the induction period, the abundance of total bacteria tended to be lower than in the control group on day 0 and was higher than in the control group on day 20 (P= 0·05 and P< 0·05, respectively). During the recovery period, the abundance of total bacteria was higher than that in the control group on days 12 and 20 (1·6-fold increase; P< 0·05).
Amylolytic bacteria
The induction diet increased starch concentration, providing more substrate for amylolytic bacteria. During the induction period, the relative abundance of S. bovis was 5-fold higher than the control levels on day 4, and remained 3·5-fold higher than the control levels throughout day 20 (P< 0·01; Fig. 2(a)) . During the recovery period, the abundance of S. bovis was higher than the control levels on day 0 (P< 0·05), but was not different from the control levels from day 4 to 20. In contrast, the abundance of P. bryantii was not different from the control levels from day 0 to 12 of induction, but was 56 % lower than the control levels on day 20 (P= 0·03; Fig. 2(b)). During the recovery period, the abundance of P. bryantii tended to be lower than the control levels on days 0 and 8 (P< 0·10) and was 6-fold lower than the control levels on day 4 (P< 0·01), but was not different on days 12 and 20.

Fig. 2 Time course of the relative abundance of amylolytic bacteria ((a) Streptococcus bovis and (b) Prevotella
bryantii) in the rumen digesta of dairy cows fed a high-fibre, low-PUFA diet (control; ![]() ), a low-fibre, high-PUFA diet resulting in milk fat depression (induction;
), a low-fibre, high-PUFA diet resulting in milk fat depression (induction; ![]() ), or a high-fibre, low-PUFA diet following induction that resolved milk fat depression (recovery;
), or a high-fibre, low-PUFA diet following induction that resolved milk fat depression (recovery; ![]() ). Abundance of the bacterial species is expressed as a percentage of total bacterial 16S ribosomal DNA. Values are least-squares means, with their largest standard errors of the mean ((a) sem 0·017 and (b) sem 0·053). Mean value for the induction group was significantly different from that of the control group: ** P< 0·05 and *** P< 0·01 (pre-planned contrasts). Mean value for the recovery group was significantly different from that of the control group: † P< 0·10, †† P< 0·05, ††† P< 0·01 (pre-planned contrasts). For S. bovis, there were significant main effects for treatment (P< 0·001) and time (P= 0·04), and the treatment × time effect was significant (P< 0·001). For P.
bryantii, there was a significant main effect of treatment (P< 0·01), but no effect for time (P= 0·18). There was a significant treatment × time effect (P< 0·01).
). Abundance of the bacterial species is expressed as a percentage of total bacterial 16S ribosomal DNA. Values are least-squares means, with their largest standard errors of the mean ((a) sem 0·017 and (b) sem 0·053). Mean value for the induction group was significantly different from that of the control group: ** P< 0·05 and *** P< 0·01 (pre-planned contrasts). Mean value for the recovery group was significantly different from that of the control group: † P< 0·10, †† P< 0·05, ††† P< 0·01 (pre-planned contrasts). For S. bovis, there were significant main effects for treatment (P< 0·001) and time (P= 0·04), and the treatment × time effect was significant (P< 0·001). For P.
bryantii, there was a significant main effect of treatment (P< 0·01), but no effect for time (P= 0·18). There was a significant treatment × time effect (P< 0·01).
Lactic acid-utilising bacteria
Increasing diet fermentability by the addition of dietary starch results in ruminal lactate production, providing substrate for lactate-utilising bacteria. During the induction period, the abundance of M. elsdenii and S. ruminantium increased progressively and was higher than in control from day 4 to 12 (P< 0·05; Fig. 3), but declined thereafter and was not different from control on day 20. The relative abundance of M. elsdenii peaked on day 12 of induction (16-fold increase), whereas that of S. ruminantium reached near the maximum level on day 8 (1·7-fold increase v. control). When dairy cows were switched to the recovery diet, the abundance of M. elsdenii declined and was not different from the control levels from day 4 to 20. The abundance of S. ruminantium did not differ from that of the control group at any time point during the recovery from MFD.

Fig. 3 Time course of the relative abundance of lactate-utilising bacteria ((a) Megasphaera elsdenii and (b) Selenomonas ruminantium) in the rumen digesta of dairy cows fed a high-fibre, low-PUFA diet (control; ![]() ), a low-fibre, high-PUFA diet resulting in milk fat depression (induction;
), a low-fibre, high-PUFA diet resulting in milk fat depression (induction; ![]() ), or a high-fibre, low-PUFA diet following induction that resolved milk fat depression (recovery;
), or a high-fibre, low-PUFA diet following induction that resolved milk fat depression (recovery; ![]() ). Abundance of the bacterial species is expressed as a percentage of total bacterial 16S rDNA. Values are least-squares means, with their largest standard errors of the mean ((a) sem 0·0050 and (b) sem 0·043). Mean value for the induction group was significantly different from that of the control group: ** P< 0·05, *** P< 0·01 (pre-planned contrasts). †† Mean value for the recovery group was significantly different from that of the control group (P< 0·05; pre-planned contrasts). For M. elsdenii and S. ruminantium, there was a significant main effect of treatment (P= 0·02 and P< 0·01, respectively), but no effect for time (P= 0·59 and P= 0·34, respectively). There was a significant treatment × time effect (P= 0·01 and P< 0·01, respectively).
). Abundance of the bacterial species is expressed as a percentage of total bacterial 16S rDNA. Values are least-squares means, with their largest standard errors of the mean ((a) sem 0·0050 and (b) sem 0·043). Mean value for the induction group was significantly different from that of the control group: ** P< 0·05, *** P< 0·01 (pre-planned contrasts). †† Mean value for the recovery group was significantly different from that of the control group (P< 0·05; pre-planned contrasts). For M. elsdenii and S. ruminantium, there was a significant main effect of treatment (P= 0·02 and P< 0·01, respectively), but no effect for time (P= 0·59 and P= 0·34, respectively). There was a significant treatment × time effect (P= 0·01 and P< 0·01, respectively).
Fibrolytic bacteria
The induction diet decreased dietary fibre content, but increased the concentrations of unsaturated FA, which are especially toxic to many fibrolytic bacteria. The abundance of R. albus did not differ. The abundance of R. albus was not different between treatments on day 0 or from day 8 to 20; however, it tended to be lower on day 4 of induction and to be higher on day 4 of recovery relative to the control period (P= 0·07 and P= 0·08, respectively; Fig. 4(a)). The abundance of F. succinogenes decreased by 98 % relative to that of the control group on day 4 of induction, and this level was maintained throughout day 20 (P< 0·01; Fig. 4(b)). Conversely, during the recovery period, the abundance of F. succinogenes was 99 % lower than that of the control group on day 0 (P< 0·01), increased progressively, did not differ from the control levels on day 4, and was fully recovered by day 12. There was no treatment × time interaction for P. ruminicola; however, there was an effect of treatment (P< 0·01; see online Supplementary Table S2). The abundance of P. ruminicola was 18 % lower than that of the control group during the induction period (P= 0·02), and tended to be 20 % higher than that of the control group during the recovery period (P= 0·06). Based on the pre-planned contrast, the abundance of P. ruminicola was found to be lower than that of the control group on day 8 of induction (P= 0·04; Fig. 4(c)), and tended to be lower on day 12 (P= 0·08). During the recovery period, the abundance of P. ruminicola did not differ from that of the control group at any time point.

Fig. 4 Time course of the relative abundance of fibrolytic bacteria ((a) Ruminococcus albus, (b) Fibrobacter succinogenes and (c) Prevotella ruminicola) in the rumen digesta of dairy cows fed a high-fibre, low-PUFA diet (control; ![]() ), a low-fibre, high-PUFA diet resulting in milk fat depression (induction;
), a low-fibre, high-PUFA diet resulting in milk fat depression (induction; ![]() ), or a high-fibre, low-PUFA diet following induction that resolved milk fat depression (recovery;
), or a high-fibre, low-PUFA diet following induction that resolved milk fat depression (recovery; ![]() ). Abundance of the bacterial species is expressed as a percentage of total bacterial 16S rDNA. Values are least-squares means, with their largest standard errors of the mean ((a) sem 0·049, (b) sem 0·359 and (c) sem 0·247). Mean value for the induction group was significantly different from that of the control group: * P< 0·10, ** P< 0·05, *** P< 0·01 (pre-planned contrasts). Mean value for the recovery group was significantly different from that of the control group: † P< 0·10, ††† P< 0·01 (pre-planned contrasts). For R. albus, there was a significant main effect of time (P= 0·02), but no effect for treatment (P= 0·24). There was a significant treatment × time effect (P< 0·01). For F. succinogenes, there was a significant main effect of treatment (P< 0·001), but no effect for time (P= 0·13). There was a significant treatment × time effect (P< 0·001). For P. ruminicola, there was a significant main effect of treatment (P< 0·01), but no effect for time (P= 0·10). There was no significant treatment × time effect (P= 0·59).
). Abundance of the bacterial species is expressed as a percentage of total bacterial 16S rDNA. Values are least-squares means, with their largest standard errors of the mean ((a) sem 0·049, (b) sem 0·359 and (c) sem 0·247). Mean value for the induction group was significantly different from that of the control group: * P< 0·10, ** P< 0·05, *** P< 0·01 (pre-planned contrasts). Mean value for the recovery group was significantly different from that of the control group: † P< 0·10, ††† P< 0·01 (pre-planned contrasts). For R. albus, there was a significant main effect of time (P= 0·02), but no effect for treatment (P= 0·24). There was a significant treatment × time effect (P< 0·01). For F. succinogenes, there was a significant main effect of treatment (P< 0·001), but no effect for time (P= 0·13). There was a significant treatment × time effect (P< 0·001). For P. ruminicola, there was a significant main effect of treatment (P< 0·01), but no effect for time (P= 0·10). There was no significant treatment × time effect (P= 0·59).
Trans-11 18 : 1-producing bacteria
The abundance of the B. fibrisolvens/Pseudobutyrivibrio group and B. hungatei were investigated in the present study because of their role in ruminal BH of cis-9, cis-12 18 : 2 (linoleic acid; LA) to cis-9, trans-11-CLA and trans-11 18 : 1( Reference Lourenço, Ramos-Morales and Wallace 12 ). The abundance of the B. fibrisolvens/Pseudobutyrivibrio group decreased progressively during the induction period and reached a nadir on day 8 when it was 60 % lower than that of the control group (P< 0·05; Fig. 5(a)). There was no treatment × time interaction or treatment effect at any time point for the abundance of B. hungatei (see online Supplementary Table S2; Fig. 5(b)).

Fig. 5 Time course of the relative abundance of trans-11 18 : 1-producing bacteria ((a) Butyrivibrio fibrisolvens/Pseudobutyrivibrio group and (b) Butyrivibrio hungatei) in the rumen digesta of dairy cows fed a high-fibre, low-PUFA diet (control; ![]() ), a low-fibre, high-PUFA diet resulting in milk fat depression (induction;
), a low-fibre, high-PUFA diet resulting in milk fat depression (induction; ![]() ), or a high-fibre, low-PUFA diet following induction that resolved milk fat depression (recovery;
), or a high-fibre, low-PUFA diet following induction that resolved milk fat depression (recovery; ![]() ). Abundance of the bacterial species is expressed as a percentage of total bacterial 16S rDNA. Values are least-squares means, with their largest standard errors of the mean ((a) sem 0·273 and (b) sem 0·017). Mean value for the induction group was significantly different from that of the control group: ** P< 0·05, *** P< 0·01 (pre-planned contrasts). Mean value for the recovery group was significantly different from that of the control group: †† P< 0·05, ††† P< 0·01 (pre-planned contrasts). For the B. fibrisolvens/Pseudobutyrivibrio group, there was a significant effect of treatment (P< 0·01), but no effect for time (P= 0·15). There was a significant treatment × time effect (P< 0·001). For B. hungatei, there was no significant effect for treatment (P= 0·52) or time (P= 0·33), and no significant treatment × time effect (P= 0·67).
). Abundance of the bacterial species is expressed as a percentage of total bacterial 16S rDNA. Values are least-squares means, with their largest standard errors of the mean ((a) sem 0·273 and (b) sem 0·017). Mean value for the induction group was significantly different from that of the control group: ** P< 0·05, *** P< 0·01 (pre-planned contrasts). Mean value for the recovery group was significantly different from that of the control group: †† P< 0·05, ††† P< 0·01 (pre-planned contrasts). For the B. fibrisolvens/Pseudobutyrivibrio group, there was a significant effect of treatment (P< 0·01), but no effect for time (P= 0·15). There was a significant treatment × time effect (P< 0·001). For B. hungatei, there was no significant effect for treatment (P= 0·52) or time (P= 0·33), and no significant treatment × time effect (P= 0·67).
Associations between milk fat concentration, milk fatty acid profile and microbial taxa
Quadratic relationships were observed between milk fat concentration and anaerobic fungi, F. succinogenes and the B. fibrisolvens/Pseudobutyrivibrio group (R 2≥ 0·52; Table 2), whereas trans-10 18 : 1 concentration (% of C18 FA) was quadratically associated with milk fat concentration, ciliate protozoa, anaerobic fungi, F. succinogenes and the B. fibrisolvens/Pseudobutyrivibrio group (R 2≥ 0·45). There was a modest quadratic relationship between trans-11 18 : 1 (% of C18 FA) and milk fat (R 2 0·34), but trans-11 18 : 1 was not strongly associated with any microbial taxa (R 2< 0·13; see online Supplementary Table S3). The relationship between microbial populations was also investigated by regression analysis (see online Supplementary Table S3). Briefly, anaerobic fungi were correlated with protozoa (R 2 0·57) and F. succinogenes (R 2 0·76). Also, modest correlations were observed between protozoa and F. succinogenes and between S. ruminantium and the B. fibrisolvens/Pseudobutyrivibrio group (R 2>0·33).
Table 2 Associations between milk fat and trans-10 18 : 1 concentrations and selected rumen microbes

RMSE, root mean squared error; Int, intercept; L, linear coefficient; Q, quadratic coefficient; FA, fatty acid.
* Significance of the intercept, linear and quadratic terms. The quadratic effect was removed from the model when P>0·05 only if fit improved based on the Akaike information criterion.
† Predictors are expressed as a percentage of total bacteria.
Discussion
Previous investigations have predominantly focused on characterising the gastrointestinal microbiome after adaptation to a diet rather than on temporal events occurring during adaptation( Reference Belenguer, Toral and Frutos 13 – Reference Castro-Carrera, Toral and Frutos 15 ), although Tajima et al. ( Reference Tajima, Aminov and Nagamine 5 ) reported an experiment with a low-resolution time course (i.e. days 0, 3 and 28) on the changes in select bacterial groups after switching cows from a high-fibre diet to a high-grain diet. The repeated-measures design adopted in the present experiment accounted for the variation in cow and period, and strengthens the ability to investigate the time course of adaptation. To understand the variation due to cow and period, we ran the model using the sum-of-squares estimation (type 3) without a repeated statement. On average, period was 4·8 % of the sum of squares and cow was 16·5 % of the sum of squares. Cow variability was 0·8–38 % of the total sum of squares depending on the microbial population. Previous work reporting milk fat and milk FA profile every other day when cows were switched to a high-fat diet( Reference Shingfield, Reynolds and Hervas 2 ) provided insight into temporal changes in rumen metabolism and thus the expected changes in ruminal microbial populations. Samples were collected to represent different periods of the trans isomer response including the peak concentration of trans-11 18 : 1 (day 4), with trans-10 18 : 1 becoming the predominant trans isomer (days 8 and 12), and established MFD (day 20). An additional sample was collected on day 16, but was not analysed because preliminary analysis of one cow indicated insignificant difference between days 12, 16 and 20. Thus, the measurements of time course in the present study help in the mechanistic interpretation of microbial adaptation associated with MFD and modification of BH pathways. Importantly, this time course allows the separation of primary responses from secondary adaptations.
Dietary components have an impact on the ruminal environment and predominant microbial populations, which, in turn, affect the rate and pathways of BH( Reference Lourenço, Ramos-Morales and Wallace 12 , Reference Jenkins, Wallace and Moate 16 ). The induction diet was designed to cause MFD using dietary conditions that commonly occur on farms. The experimental design was successful in the induction of and recovery from MFD within the experimental periods. Although not measured in the present study, ruminal pH was expected to have been lower during the induction period due to increased dietary starch and decreased forage NDF. Decreased ruminal pH results in changes in the microbial population (specifically cellulolytic bacteria) and BH pathways, resulting in the increased formation of trans-10 18 : 1 and trans-10, trans-12-CLA( Reference Fuentes, Calsamiglia and Cardozo 17 ). The induction diet also had a high concentration of rapidly available PUFA, specifically LA (3·4 % DM), which is known to have a negative impact on bacterial membrane integrity and the growth of numerous species of ruminal bacteria( Reference Maia, Chaudhary and Figueres 18 , Reference Maia, Chaudhary and Bestwick 19 ). Increasing PUFA also promotes a shift in microbial communities and BH pathways towards those that cause MFD( Reference Weimer, Stevenson and Mertens 4 , Reference Lourenço, Ramos-Morales and Wallace 12 ). Given that the bacterial species assayed in the present study represent a small proportion of the total community, cause-and-effect relationships cannot be drawn from the present study. However, we propose that the timing of changes in all the identified taxa, including protozoa and fungi, provide an indication of the timing of the global adaptation of the rumen microbiome previously reported to occur during MFD( Reference Weimer, Stevenson and Mertens 4 ).
Ciliate protozoa are the largest and most important group of protozoa in the rumen( Reference Williams, Coleman, Hobson and Stewart 20 ), and anaerobic fungi (order Neocallimastigales) encompasses several families and species with diverse activities( Reference Hibbett, Binder and Bischoff 21 ). Both ciliate protozoa and fungi were acutely responsive to the induction and recovery diets, and demonstrated the rapid adaptation of the rumen to the change in the diet. The regression analysis revealed that milk fat concentration quadratically increased with increasing number of protozoa, peaked when protozoa were approximately 6 % of total bacteria, and then decreased with a greater number of protozoa; whereas milk fat trans-10 18 : 1 concentration quadratically decreased with increasing number of protozoa, and reached a nadir when the number of protozoa was about 4 % of total bacteria (Table 2). Rumen ciliate protozoa are known to contain high concentrations of trans-11 18 : 1 and CLA( Reference Yáñez-Ruiz, Scollan and Merry 22 , Reference Devillard, McIntosh and Newbold 23 ); however, they do not directly contribute to the BH of LA( Reference Devillard, McIntosh and Newbold 23 ) and were not correlated with the concentration of trans-11 18 : 1 in milk fat in the present study (R 2 0·08; P= 0·09; see online Supplementary Table S3).
It is unclear whether the rapid reduction in the abundance of ciliate protozoa and fungi during the induction of MFD in the present study is due to the high-PUFA diet, high diet fermentability, or their interaction. The reported effects of high-grain diets and low ruminal pH on rumen protozoa are contradictory. Low ruminal pH under high-grain diet conditions has been previously shown to have a negative impact on the abundance of rumen protozoa in vitro ( Reference Dehority 24 ) and in vivo ( Reference Franzolin and Dehority 25 , Reference Khafipour, Li and Plaizier 26 ). In contrast, other studies have observed positive effects of high-grain diets on the abundance of protozoa( Reference Franzolin and Dehority 27 , Reference Hook, Steele and Northwood 28 ). Dietary PUFA have been reported to be toxic to protozoa populations, although the degree of inhibition is greater for linseed than for other oils such as soyabean and rapeseed( Reference Doreau and Ferlay 29 ). In agreement with our observations under the MFD induction diet condition, feeding a ration containing sunflower oil (6 % DM) decreased ciliate protozoa concentrations in ruminal fluid by 80 % within 6 d of feeding( Reference Ivan, Mir and Koenig 30 ).
Very few data exist on the effects of diet on ruminal fungi and their role in ruminal BH, although some species (e.g. Neocallimastix frontalis) are known to metabolise LA to trans-9, trans-11-CLA, whereas the growth of others (e.g. Piromyces communis) is inhibited by LA( Reference Maia, Chaudhary and Figueres 18 ). Using automated ribosomal intergenic spacer analysis, Boots et al. ( Reference Boots, Lillis and Clipson 31 ) recently investigated the effects of increasing dietary concentrates with or without the addition of soyabean oil (6 % DM), and reported a more pronounced reduction in anaerobic fungal diversity in the rumen when oil was increased compared with when concentrate was increased.
The abundance of F. succinogenes, P. ruminicola, M. elsdenii, S. ruminantium and S. bovis in the control group was similar to that reported previously by Stevenson & Weimer( Reference Stevenson and Weimer 6 ), whereas the abundance of R. albus was higher and that of P. bryantii was lower. Interestingly, although M. elsdenii is considered a major lactic acid user in the rumen( Reference Counotte, Prins and Janssen 32 ), its relative abundance was much lower than that of S. ruminantium in the present experiment, similar to that reported previously( Reference Stevenson and Weimer 6 ). Although not measured, lactic acid concentration in the rumen was expected to have been increased under the MFD induction diet condition. Compared with high-forage diets, feeding high-grain diets increased the formation of lactic acid by ruminal bacteria such as S. bovis ( Reference Russell and Hino 33 , Reference Owens, Secrist and Hill 34 ), which drastically increased during the induction period. In agreement with our observations, the abundance of M. elsdenii was also elevated in milk fat-depressed cows receiving a low-forage diet containing flaked ‘corn’( Reference Latham, Storry and Sharpe 35 ), in cows fed high-grain diets( Reference Khafipour, Li and Plaizier 26 ), and in two cows with low milk fat fed a monensin-supplemented high-‘corn’ silage diet( Reference Palmonari, Stevenson and Mertens 36 ). The more pronounced increased in the abundance of M. elsdenii in the present experiment could be related to its lower sensitivity to the toxic effects of PUFA( Reference Maia, Chaudhary and Figueres 18 ). Interestingly, both species decreased to near control values by day 20 of induction, suggesting a possible secondary adaptation to the diet, which is supported by a decrease in the abundance of S. bovis after day 4, or a long-term detrimental effect of the diet on the growth of the lactate users. In addition, we observed that trans-10 18 : 1 in milk fat was quadratically related to M. elsdenii (increased with increasing abundance of M. elsdenii and peaked when M. elsdenii was approximately 0·7 % of total bacteria), whereas it increased linearly with increasing abundance of S. ruminantium. The YJ4 strain of M. elsdenii has been reported to produce trans-10, trans-12-CLA from linoleic acid( Reference Kim, Liu and Rychlik 37 ), although others have reported that other strains do not possess this ability( Reference Wallace, Chaudhary and McKain 38 ). However, Propionibacterium acnes has been suggested to be a more likely candidate for ruminal synthesis of trans-10, cis-12-CLA( Reference Lourenço, Ramos-Morales and Wallace 12 ), and other unculturable species probably are major contributors.
When transitioning cows from a high-hay to a high-grain diet, Tajima et al. ( Reference Tajima, Aminov and Nagamine 5 ) observed a sharp initial increase in the abundance of the amylolytic bacteria S. bovis and P. bryantii on day 3 and a large reduction in the number of both bacterial species by day 28. In contrast, in the present experiment, S. bovis and P. bryantii responded differently during the induction period. While the two bacterial species degrade starch avidly( Reference Cotta 39 , Reference Chiquette, Allison and Rasmussen 40 ), P. bryantii also exhibits xylanase and pectinase activities( Reference Stewart, Flint, Bryant, Hobson and Stewart 41 ) and appeared to be part of the second phase of rumen adaptation, whereas S. bovis appeared to be part of the first phase. This difference in response may be explained by the higher sensitivity of P. bryantii to PUFA compared with S. bovis ( Reference Maia, Chaudhary and Figueres 18 ). Temporally, the peak abundance of S. bovis coincided with peak milk concentration of vaccenic acid, and the decline in the abundance of S. bovis after day 4 coincided with the shift to the alternative BH pathway( Reference Rico and Harvatine 3 ). However, S. bovis was poorly correlated (R 2< 0·30) with milk trans-11 18 : 1 concentration, but was correlated with milk fat concentration (quadratic and increased with the abundance of S. bovis above 0·08 % of total bacteria) and trans-10 18 : 1 concentration (quadratic and decreased with the abundance of S. bovis above 0·1 % of total bacteria). Although S. bovis is not known to be important in the ruminal formation of trans-11 18 : 1 or CLA( Reference Maia, Chaudhary and Figueres 18 ), it is known to biohydrogenate MUFA and PUFA( Reference Hudson, Morvan and Joblin 42 ). Interestingly, the decline in the abundance of P. bryantii during the induction period coincided with the alternative BH pathway that became the predominant pathway, and its increase during the induction period coincided with the decline in the alternative pathway and re-establishment of the normal pathway. However, it was poorly correlated with the concentrations of milk trans-10 18 : 1 and milk fat.
The fibrolytic species of bacteria selected in the present experiment also differed in their response to dietary changes possibly due to differences in sensitivity to ruminal pH and PUFA. In agreement with the present results, Tajima et al. ( Reference Tajima, Aminov and Nagamine 5 ) reported a dramatic reduction in the abundance of F. succinogenes on day 3 of feeding a high-grain diet, which was further reduced by day 28. The rapid decline in the abundance of F. succinogenes coincided with the increase in the concentration of milk fat trans-11 18 : 1, but was poorly correlated to this FA, and further changes were not observed during the shift to the alternative pathway. The increase in the abundance of F. succinogenes during the recovery from MFD temporally matched the decline in the alternative BH pathway. In contrast, P ruminicola seemed to be less responsive to dietary changes in the present experiment as it was only mildly reduced during the induction period and was poorly associated with the concentrations of milk fat and trans-10 18 : 1. However, P. ruminicola differs from other more specialised cellulolytic bacteria as it has the ability to ferment peptides and amino acids. Interestingly, R. albus was only affected on day 4 of the induction and recovery periods, and may represent a transitory role in rumen adaptation.
Ruminal bacteria are generally considered to be the main group accounting for ruminal BH of unsaturated FA( Reference Lourenço, Ramos-Morales and Wallace 12 ). Both the B. fibrisolvens/Pseudobutyrivibrio group and B. hungatei produce trans-11 18 : 1( Reference Paillard, McKain and Chaudhary 43 , Reference Boeckaert, Boon and Verstraete 44 ), although in the present experiment, both were poorly associated with trans-11 18 : 1 (B. fibrisolvens/Pseudobutyrivibrio group R 2 0·13 and B. hungatei R 2 0·03). Despite their commonalities with regard to BH products and their phylogenetic proximity, B. hungatei is known to have higher butyrate kinase activity and to be more sensitive to growth inhibition by LA compared with the B. fibrisolvens/Pseudobutyrivibrio group( Reference Paillard, McKain and Chaudhary 43 ). The inclusion of 2 % sunflower oil in the diet of dairy ewes had no effect on the relative abundance of trans-11 18 : 1-producing Butyrivibrio. In contrast, the abundance of B. fibrisolvens/Pseudobutyrivibrio group progressively decreased during the induction of MFD in the present experiment, whereas no changes were observed in the abundance of B. hungatei. It is possible that the negative effects of the MFD induction diet on the abundance of B. fibrisolvens/Pseudobutyrivibrio group were related to the reduction in dietary fibre content, as B. fibrisolvens-related bacteria are known to be involved in fibre digestion( Reference Hespell, Wolf and Bothast 45 ). The gradual decline in the abundance of the B. fibrisolvens/Pseudobutyrivibrio group during the induction period temporally matched the shift to the alternative BH pathway, although the gradual recovery lagged behind the recovery of the normal pathway.
Conclusions
Changes in dietary fibre and unsaturated FA concentrations resulting in the induction of and recovery from milk fat depression also cause a rapid alteration of ruminal microbial populations. Only a small proportion of total bacteria were evaluated; however, the time course of rumen taxa modifications and their correlations with milk fat and milk trans-FA concentrations are expected to reflect the time course of global microbial shifts. Changes in the rumen microbiome may be the functional and rate-limiting step in the induction of and recovery from diet-induced milk fat depression; however, further investigation is needed to elucidate the functional role of specific bacterial populations in the process of modification of dietary lipids.
Supplementary material
To view supplementary material( Reference Ouwerkerk, Klieve and Forster 46 – Reference Sylvester, Karnati and Yu 49 ) for this article, please visit http://dx.doi.org/10.1017/S0007114515001865
Acknowledgements
The authors acknowledge Jackie Ying, Andrew Clark and L. W. Rottman (Penn State) for technical assistance, and the Genomics Core Center in the Huck Institutes of the Life Sciences at Penn State University for the use of equipment.
ADM Alliance Animal Nutrition (Decatur, IL) donated the Amino Plus for the present experiment. The present study was supported by the Agriculture and Food Research Initiative Competitive Grant (2010-65206-20723, to K. J. H.) from the USDA National Institute of Food and Agriculture (Washington, DC) and Penn State University (University Park, PA).
The authors' contributions were as follows: K. J. H. and D. E. R. designed the research and wrote the paper; D. E. R. conducted the animal experiment and data analysis; D. E. R., S. H. P. and J. M. R. conducted the microbial analysis with guidance from K. J. H.; K. J. H. had primary responsibility for the final content. All authors read and approved the final manuscript.
The authors declare that they have no conflicts of interest.










