Perilipin is a protein localised on lipid droplet surfaces in adipocytes and steroidogenic cells(Reference Blanchette-Mackie, Dwyer and Barber1), and plays a central role in lipid storage and breakdown. At a basal state, perilipin protects the lipid core from lipases (e.g. hormone-sensitive lipase and adipocyte TAG lipase) by sequestering the enzymes from the lipid droplet surfaces(Reference Marcinkiewicz, Gauthier and Garcia2). However, once phosphorylated, perilipin allows or even recruits lipases to access lipid droplets(Reference Marcinkiewicz, Gauthier and Garcia2), and hence causes active lipolysis.
Several studies have shown that common polymorphisms at the PLIN1 gene are associated with obesity risk(Reference Qi, Corella and Sorli3–Reference Corella, Qi and Sorli6) and insulin resistance(Reference Tai and Ordovas7). Specifically, the PLIN1 11482G>A polymorphism was associated with weight-loss resistance in obese patients(Reference Corella, Qi and Sorli6). Others have also reported significant gene–diet interactions with several dietary components(Reference Corella, Qi and Tai8, Reference Smith, Tucker and Yiannakouris9). More recently, Jenkins et al. (Reference Jenkins, McKenzie and Damcott10) have shown a PLIN1–exercise interaction with obesity and several metabolic factors.
Studies examining the effects of PLIN1 polymorphisms on energy and substrate utilisation rate changes after an energy restriction treatment in obese patients are, however, scarce. Obesity treatment strategies could be improved if predictive information about the response to therapy were available.
We investigated the association of PLIN1 11482G>A (rs894160) and PLIN1 13041A>G (rs2304795) polymorphisms with body composition, energy and substrate metabolism, and the metabolic response to a 12-week energy-restricted diet in obese women.
Methods
Study population
A total of eighty-three obese women (BMI inclusion criteria 30–39·9 kg/m2) from Vitoria (North Spain) aged between 19 and 49 years volunteered to participate in the present study and underwent a comprehensive medical examination. Participants were premenopausal, non-athletic and showed weight stability (body weight changes < 3 kg in the last 3 months). Exclusion criteria included history of CVD or diabetes, pregnancy, total cholesterol levels >3000 mg/l, TAG levels >3000 mg/l and blood pressure level >140/90 mmHg. We also excluded women under medication (except oral contraceptives) for hypertension, hyperlipidaemia, hyperuricaemia or other illness.
Of the eighty-three participants, four left the study due to inability to follow the research protocol and one left due to pregnancy. Only the data from women who finished the 12-week diet intervention programme, i.e. n 78, were included in the analyses (dropout rate 6 %). The final sample did not differ in key characteristics (i.e. age, BMI or total body fat mass (FM)) from the original sample (all P>0·1).
All women received verbal and written information about the nature and purpose of the survey, and all of them gave written consent for participation in the study. The present study was in accordance with the Helsinki II Declaration and was approved by the Ethical Committee in Hospital of Txagorritxu (Vitoria, Spain).
Design
The present study was designed as a 12-week controlled body-weight-loss programme. Body weight reduction (0·5–1 kg/week) was induced by a low-energy mixed (55 % carbohydrate, 30 % lipid and 15 % protein) diet providing 2510 kJ (600 kcal) less than individually estimated energy requirements based on measured RMR and multiplied by a factor of 1·3, as corresponds to a low physical activity level. Energy content and macronutrient composition of diets were according to the American Diabetes Association nutrition recommendations(Reference Bantle, Wylie-Rosett and Albright11).
Measurements were performed before and after a 12-week energy-restricted treatment in the Unit of Clinic Assays of LEIA Foundation (Txagorritxu Hospital). Fasting blood samples were taken from an antecubital vein after gas exchange measurement (Vmax; Sensormedics, Hochberg, Germany). Samples were processed after collection and stored at − 80°C for later analysis. Body weight ( ± 10 g) was measured after voiding using a digital integrating scale (SECA 760; SECA, Hamburg, Germany). Height was measured to the nearest 5 mm using a stadiometer (SECA 220; SECA) at the start of the study. BMI was calculated as weight (kg)/height (m2).
Body composition
Dual-energy X-ray absorptiometry measurements were performed at pre- and post-intervention examinations. A dual-energy X-ray absorptiometry scanner 140 (Hologic, QDR 4500W; Bedford, MA, USA) with QDR software for windows version 12.4 was used to estimate FM, bone-free lean tissue mass (LM) and bone mass (BM). Percentage of FM was calculated as (FM/(FM+LM+BM) × 100).
Indirect calorimetry
Fasting RMR and non-protein respiratory quotient were estimated by respiratory exchange measurements, as described elsewhere(Reference Labayen, Forga and Martinez12, Reference Labayen, Diez and Gonzalez13). Fasting urine was collected (between approximately 21.00–22.00 hours of the day before the examination and 08.00–09.00 hours of the examination day) to determine nitrogen output; glucose and lipid oxidation rates were thereafter calculated(Reference Labayen, Diez and Parra14).
Biochemical assays
Blood samples were taken following a 12 h overnight fast. Fasting plasma glucose (mmol/l) and cholesterol (mmol/l) were measured by an enzymatic spectrophotometric technique with an autoanalyser (COBAS FARA; Roche Diagnostics, Basel, Switzerland). Insulin (mIU/l) and leptin (ng/ml) were measured by ELISA kits (Linco Research, St Charles, MO, USA). Insulin sensitivity was assessed by the homeostasis model of assessment for insulin resistance. All samples were measured in duplicate, and the mean was scored.
Genotyping
Genomic DNA was isolated from the buffy coat of centrifuged whole blood using the QIAamp DNA Blood Mini Kit (Qiagen, Dusseldorf, Germany) according to the manufacturer's instructions. At the PLIN1 locus, two polymorphisms were genotyped: PLIN1 11482G>A (rs894160) and PLIN1 13041A>G (rs2304795). Genotyping was carried out using TaqMan probes and an Applied Biosystems 7300 Sequence Detection System (Applied Biosystems, Foster City, CA, USA). The final success rate for PLIN1 genotyping in the study participants was 100 %, and no discordant genotypes were observed in duplicate samples.
Statistical analysis
Hardy–Weinberg equilibrium was tested using Pearson's χ2 test for each polymorphism.
We analysed mean differences of the study variables across the PLIN1 11482G>A and 13041A>G genotypes using ANCOVA, adjusting for age. Likewise, differences in changes (after a 12-week energy-restricted diet) in the outcome variables across the PLIN1 11482G>A and 13041A>G genotypes were analysed using ANCOVA adjusting for age. We tested the additive model, and when evidence of an association was found, we grouped carriers of the less common alleles and compared with the wild-type genotypes (dominant model)(Reference Corella, Qi and Sorli6). To test for the existence of an interaction effect between the studied polymorphisms and diet-induced changes on the outcome variables, we used the ANCOVA for repeated measures, and we added a cross-product term polymorphism × diet-induced changes into the model. Percentage of change in the outcome variables after the dietary energy-restricted diet was calculated as: Δ (%): ((baseline − post-intervention)/baseline) × 100. Analyses were performed using the Statistical Package for Social Sciences (version 17.0 for Windows; SPSS, Inc., Chicago, IL, USA), and the level of significance was set to 0·05, except for the interaction effect that was considered to be 0·1.
Results
Characteristics of the study sample before and after the 12-week energy-restricted diet intervention in obese women are presented in Table 1. Genotype distributions did not deviate from Hardy–Weinberg expectations for the two PLIN1 polymorphisms. Genotype frequencies were 32 (41·0 %), 36 (46·2 %) and 10 (12·8 %) for the GG, GA and AA genotypes of the PLIN1 11482G>A polymorphism, respectively (Table 2) and 26 (33·3 %), 39 (50 %) and 13 (16·7 %) for the AA, AG and GG genotypes of the PLIN1 13041A>G polymorphism, respectively.
Table 1 Characteristics of the study sample at baseline and after the 12-week energy-restricted diet intervention
(Mean values and standard deviations)
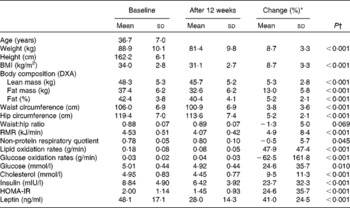
DXA, dual-energy X-ray absorptiometry; HOMA-IR, homeostasis model of assessment for insulin resistance.
* Change (%): ((baseline − post-intervention)/baseline) × 100.
† Age-adjusted from repeated-measures ANCOVA.
Table 2 Body composition, energy metabolism and biochemical variable changes* after the 12-week energy-restricted diet intervention according to the PLIN1 11482G>A (rs894160) genotypes in obese women†
(Mean values and standard deviations)
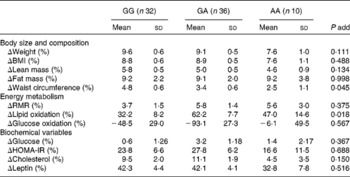
add, Additive; HOMA-IR, homeostasis model of assessment for insulin resistance.
* % Change calculated as: ((baseline − post-intervention)/baseline) × 100.
† Variables with a skewed distribution were logarithmically transformed, i.e. plasma cholesterol, insulin, leptin and glucose and lipid oxidation rates to obtain a more symmetrical distribution.
There was an interaction effect between the PLIN1 11482G>A polymorphism and diet on waist circumference (P = 0·064) and lipid oxidation rate (P = 0·004). Indeed, the minor A allele of the PLIN1 11482G>A polymorphism was associated with a lower waist-circumference change ( − 1·2 % per risk allele (95 % CI − 2·4, − 0·03), age-adjusted P for trend = 0·045). The result did not substantially change after further adjustment for height and FM change (P for trend = 0·039). Women carrying the minor A allele of the PLIN1 11482G>A polymorphism had a lower reduction in waist circumference than non-A allele carriers (3·2 (sd 0·5) v. 4·6 (sd 0·6) %, respectively, P = 0·047).
The minor A allele of the PLIN1 11482G>A polymorphism was also associated with a higher decrease in lipid oxidation rates (13·6 % per risk allele (95 % CI − 2·0, 29·0), age-adjusted P for trend = 0·018). The findings did not substantially change after further adjusting for weight-loss change (P = 0·011). Moreover, women with the minor A allele of the PLIN1 11482G>A polymorphism had a higher decrease in lipid oxidation rate than non-A allele carriers (58·9 (sd 6·7) v. 31·3 (sd 8·2) %, respectively, P = 0·012).
We observed no significant interaction effect between the PLIN1 13041A>G polymorphism and diet on body composition, energy metabolism or biochemical variables (data not shown).
Discussion
The present study shows a possible effect of the PLIN1 11482G>A polymorphism in modulating metabolic changes induced by an energy-restricted diet intervention. Obese women with the minor A allele had a lower decrease in abdominal adiposity independently of total FM loss and a higher decrease in lipid oxidation rates regardless of total body-weight loss after the 12-week energy-restricted diet intervention. Conversely, we found no effect of the PLIN1 13041A>G polymorphism on diet-induced changes, which concurs with other studies(Reference Qi, Tai and Tan5, Reference Jang, Kim and Lee15, Reference Deram, Nicolau and Perez-Martinez16).
The PLIN1 11482G>A polymorphism has been associated with resistance to weight loss in women(Reference Corella, Qi and Sorli6). Perilipin inhibits basal lipolysis and promotes TAG storage by limiting lipase access to TAG stores but also participates in catecholamine-stimulated lipolysis through an interaction with lipid droplet-associated hormone-sensitive lipase(Reference Marcinkiewicz, Gauthier and Garcia2). PLIN genetic variants affect the protein content and lipolytic rates of adipocytes(Reference Tai and Ordovas7). Mottagui-Tabar et al. (Reference Mottagui-Tabar, Ryden and Lofgren17) reported that the PLIN1 11482A allele was associated with a decreased perilipin content and with increased basal and noradrenaline-induced lipolysis. They also observed that the low perilipin content was associated with high in vivo lipolytic activity. These results are in agreement with studies reporting a lower obesity risk(Reference Qi, Corella and Sorli3–Reference Corella, Qi and Sorli6) and metabolic syndrome risk(Reference Tai and Ordovas7, Reference Corella, Qi and Tai8) in women. In contrast, others have observed no association of several PLIN polymorphisms (1234C>G, 3625A>G and 10438T>C) with BMI and waist circumference, nor associated with 3-year changes in these indicators in a German population with a family history of the metabolic syndrome or type 2 diabetes(Reference Bergmann, Li and Reimann18).
The fact that carriers of the A allele might have higher difficulties in reducing body weight(Reference Corella, Qi and Sorli6) or central body fat (as observed in the present study) in response to an energy-restricted diet might be partially explained by a reduced lipid oxidation rate. Indeed, we observed that changes in lipid oxidation rate were significantly associated with changes in FM measured by dual-energy X-ray absorptiometry (age-adjusted r 0·730; P = 0·013). This is of interest, since it is known that individuals with lower lipid oxidation rates after weight loss are those who are most susceptible to weight regain(Reference Labayen, Diez and Gonzalez13). Changes in fat oxidation in response to the same degree of short-term negative energy balance and long-term change in body weight are highly variable between individuals, and candidate genes for fat oxidation are likely to play an important role(Reference Corpeleijn, Petersen and Holst19).
Due to the relatively small sample size of the present study, these findings should be taken as preliminary. Further studies with bigger sample sizes in other well-characterised populations, and in men, where the effect of these polymorphisms seems to be different(Reference Qi, Corella and Sorli3, Reference Qi, Shen and Larson4, Reference Jenkins, McKenzie and Damcott10) are warranted. It should be kept in mind that the effects reported in the present study would become statistically non-significant after correction for multiple testing. However, to conclude negatively from a purely statistical point of view would be too stringent.
We believe that the modulating effects of the PLIN1 13041A>G polymorphism are informative and clinically relevant and may help in future prevention strategies of type 2 diabetes(Reference Lindstrom, Neumann and Sheppard20), obesity and related metabolic disorders(Reference Tai and Ordovas7). The great variability to the response to a diet in humans suggests that genes play a key role. Definition of the modulating effects of candidate genes for obesity will allow the identification of individuals likely to be resistant to diet or physical activity interventions(Reference Ordovas and Smith21), as well as the identification of those at risk of the development of obesity and related metabolic disorders(Reference Tai and Ordovas7). These results provide further insights into the potential modulating effect of certain genotypes on weight reduction treatments, and contribute to the body of knowledge regarding the potential benefits of personalised medicine.
Acknowledgements
We thank the women for their participation in the study and Lurdes Barrenetxea; Silvia Francisco, Izaskun Felipe and Emilio Sanz for their contribution to the subject's recruitment and medical supervision of the study. The present study was supported by the University of the Basque Country (UPV 05/80), Social Foundation of the Caja Vital-Kutxa, by the Department of Health of the Government of the Basque Country (2008/111062) and by the Spanish Ministry of Science and Innovation (RYC-2010-05957). J. R. R. and I. L. wrote the manuscript and performed the statistical analysis; J. R. R., E. L., J. M., R. A., P. A. and I. L. contributed to the interpretation and discussion of the results, and critically revised the drafted manuscript. None of the authors had a personal or financial conflict of interest.




