Maternal dietary protein restriction is known to affect fetal development(Reference Jahan-Mihan, Rodriguez and Christie1,Reference Wu, Bazer and Cudd2) profoundly. Observational and experimental studies have shown that fetal growth is most vulnerable to maternal diets during the first trimester of gestation(Reference Jahan-Mihan, Rodriguez and Christie1,Reference Wu, Bazer and Cudd2,Reference Fernandez-Twinn, Ozanne and Ekizoglou3,Reference Morrison and Regnault4) . The gestational protein restriction rat model, with 5–10 % protein compared with the 20 % protein in the control diet, has been one of the most widely researched models(Reference Zohdi, Lim and Pearson5). Gestational low-protein (LP) diets can impair fetal development by programming metabolic response pathways and regulating the expression of stress-response genes in offspring and may lead to long-term physiological consequences. For instance, a gestational LP diet could alter the expression of a set of hepatic genes and induced an altered metabolic phenotype in the liver of offspring(Reference Lillycrop, Phillips and Jackson6,Reference Burdge, Slater-Jefferies and Torrens7) , which may provide a mechanism for the impaired lipid and carbohydrate metabolism induced by maternal protein restriction observed in later life(Reference Torres, Bautista and Tovar8,Reference Zheng, Xiao and Zhang9) . Investigating these metabolic pathways and stress-induced genes in the offspring’s response to the maternal LP environment could improve the understanding of their lifelong phenotypic changes.
Dietary protein restriction is associated with numerous metabolic responses, including the activation of the eukaryotic translation initiation factor 2 (eIF2a)/activating transcription factor 4 (ATF4) pathway(Reference Harding, Novoa and Zhang10). This pathway has been well characterised in cell culture models by amino acid limitation(Reference B’Chir, Maurin and Carraro11,Reference Kilberg, Shan and Su12,Reference Bruhat, Jousse and Wang13) . The initial steps are the accumulation of tRNA and the phosphorylation of serine 51 of the α subunit of eIF2a by GCN2 kinase. Then, the increased phosphorylation of eIF2α inhibits general protein synthesis but activates the translation of ATF4. ATF4, a master regulator of stress-induced genes, plays a key role in adaptation to nutrient stress by regulating the transcription of these genes. For instance, the expression of Atf3 and Chop can be induced by ATF4, thus upregulating the amino acid response (AAR) pathway, which in turn affects the protein metabolism that helps to adapt to the LP environment(Reference Zhou and Pan14). Recent studies have shown that the AAR pathway can also be triggered in response to a maternal LP diet in dams, placenta and pups(Reference Strakovsky, Zhou and Pan15,Reference Wang, Wilson and Zhou16) . Strakovsky et al. showed that a gestational LP diet could activate the eIF2a/ATF4 pathway and further increase the expression of AAR downstream genes such as Atf3 and Chop in the placenta(Reference Strakovsky, Zhou and Pan15). Moreover, the activation of the AAR pathway has been found to be induced in the skeletal muscles of both dams and pups by a gestational LP diet(Reference Wang, Wilson and Zhou16). These studies suggest that the eIF2a/ATF4 pathway can play an essential role in adapting to maternal protein restriction by programming the AAR pathway. However, more metabolic pathways might also be involved and need to be discovered.
The eIF2a/ATF4 pathway regulates stress-induced responses during maternal protein restriction. Among stress responses, autophagy pathways have attracted extensive interest. Autophagy is a cellular process that degrades cytoplasmic proteins and organelles via lysosomal pathways upon nutrient stress(Reference Mizushima and Komatsu17,Reference Maiuri, Zalckvar and Kimchi18) . By recycling amino acids and other nutrients from soluble proteins and partial organelles in the cytoplasm, autophagy is involved in protein metabolism and basic metabolic processes to maintain normal energy levels and intracellular homoeostasis(Reference Mizushima and Komatsu17). Recent studies have found that activated eIF2a/ATF4 up-regulates autophagy-related gene expression. B’Chir et al. revealed that the activation of the eIF2a/ATF4 pathway is required for autophagy-related gene expression in amino acid-starved cells(Reference B’Chir, Maurin and Carraro11). Amino acid deprivation induces eIF2α phosphorylation and activates the expression of ATF4 and C/EBP homology protein (CHOP), which increases the transcription of a set of autophagy-related genes. These activated genes may contribute to the formation, maturation and function of autophagosomes(Reference B’Chir, Maurin and Carraro11). Wang et al. were the first to report that a gestational LP diet induces autophagy-related gene expression in the skeletal muscles of rat offspring(Reference Wang, Wilson and Zhou16). However, the role of eIF2a/ATF4 pathway in the increased expression of autophagy-related genes in the offspring and the specific target genes of ATF4 and CHOP are not fully understood.
The present study investigated hepatic autophagy-related gene expression regulated by the eIF2α/ATF4 pathway in both male and female offspring of dams fed a gestational LP diet. The hypothesis states that maternal protein restriction during gestation will induce metabolic stress responses in the liver of offspring, which occurs with a transcriptional programme of autophagy-related gene expression in a sex-specific manner to adapt to the nutrient-deprivation stress.
Methods
Animal treatments
Timed-pregnant Sprague-Dawley rats were purchased from Charles River Laboratories and were individually housed in standard polycarbonate cages in a humidity- and temperature-controlled room with free access to food and water on a 12-h light/12-h dark cycle. They were fed either a control diet (C group, 18 % energy from protein) or a LP diet (LP group, 9 % energy from protein) (Bioserve) from gestation day 2 during gestation. The experimental diets were designed according to well-established diet formula(Reference Lillycrop, Phillips and Jackson6,Reference Zhou and Pan14,Reference Zheng, Rollet and Pan19) as listed in Table 1. All dams were fed with the same control diet after delivery during lactation. After weaning, all pups were kept on the same control diet until sacrifice at postnatal day 38 (Fig. 1). One pup per dam was used for further experiments; therefore, our experimental unit was a single animal (n 8 for each group). All pups were individually housed in standard polycarbonate cages in a humidity- and temperature-controlled room with free access to food and water on a 12-h light/12-h dark cycle. The pups were killed by CO2 asphyxiation at postnatal day 38 following 12 h of fasting. The liver from each pup was collected, snap-frozen in liquid N2 and stored at –80°C until it can be processed. The animal protocol for this study was approved by the International Animal Care and Use Committee at the University of Illinois.
Table 1. Nutrient composition of the experimental diets
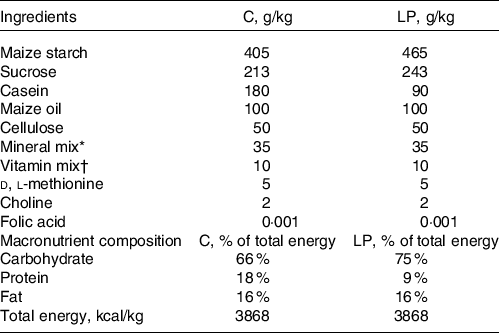
* Mineral mix: calcium phosphate dibasic 11·3 g/kg; sodium chloride 1·7 g/kg; potassium citrate monohydrate 5·0 g/kg; potassium sulphate 1·2 g/kg; magnesium sulphate 0·5 g/kg; magnesium carbonate 0·1 g/kg; ferric citrate 0·1 g/kg; zinc carbonate 36·2 mg/kg; cupric carbonate 6·8 mg/kg; potassium iodate 0·2 mg/kg; sodium selenite 0·2 mg/kg; chromium potassium sulphate 12·5 mg/kg.
† Vitamin mix: thiamine hydrochloride 2·4 mg/kg; riboflavin 2·4 mg/kg; pyridoxine hydrochloride 2·8 mg/kg; nicotinic acid 12·0 mg/kg; d-calcium pantothenate 6·4 mg/kg; biotin 0·01 mg/kg; cyanocobalbumin 0·003 mg/kg; retinyl palmitate 6·4 mg/kg; dl-μ-tocopherol acetate 79·9 mg/kg; cholecalciferol 1·0 g/kg; menaquinone 0·02 mg/kg.
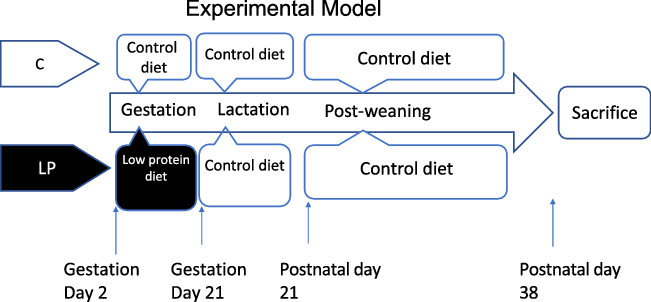
Fig. 1. The experimental design of animal study. C, pregnant rat dams fed on a control diet; LP, pregnant rat dams fed on a low-protein diet.
RNA isolation and real-time quantitative PCR
Liver tissue samples were ground in liquid N2 in a mortar and pestle, and total RNA was isolated using TRIZOL reagent (Sigma-Aldrich) as described previously(Reference Wang, Wilson and Zhou16). Total mRNA was used for DNA synthesis using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems), which was performed in a thermocycler (Applied Biosystems) at 25°C for 10 min, 37°C for 2 h and 85°C for 5 s. PCR was performed with a 96-well plate using a SYBR Green PCR Master mix (Applied Biosystems) on the StepOnePlus Real-Time PCR System (Applied Biosystems). The reaction conditions were 95°C for 15 min, 95°C for 15 s and 60°C for 60 s for forty cycles. Table 2 lists the primer sequences and source for each primer used in this study. After quantification, the ribosomal protein L7a was used to normalise all the mRNA data.
Table 2. List of primers
Protein isolation and Western blotting
Protein concentrations were determined by the Lowry assay. Western blotting was performed as described by Wang et al. (Reference Wang, Wilson and Zhou16). Frozen liver tissue (50 mg) from offspring was ground and suspended in 400 μl of protein sample buffer with a 1X proteinase inhibitor (Roche Applied Science) and phosphatase inhibitor cocktails 1 and 2 (Sigma-Aldrich). Each diluted sample (30 μg) was size-fractionated by 12 % SDS-PAGE, transferred to a 0·2-μm PVDF membrane (Bio-Rad) and blocked in 10 % non-fat dry milk in Tris Buffered Saline with Tween for 1 h at room temperature. Antibodies against target proteins were incubated with the membrane in 10 % non-fat dry milk at a 1:1000 dilution at room temperature for 1 h. Blots were incubated in Super Signal West Dura Extended Duration Substrate (Pierce) for 5 min, and then the signals were detected and analysed by Bio-Rad ChemiDoc and Quantity One software. The antibodies are listed in Table 3.
Table 3. List of antibodies
Chromatin immunoprecipitation
To investigate the roles of the transcription factors ATF4 and CHOP in regulating autophagy-related gene expression, chromatin immunoprecipitation analysis was performed according to a modified protocol(Reference Chen, Pan and Dudenhausen20) with magnetic beads as published previously(Reference Strakovsky, Wang and Engeseth21) for this study. Briefly, 200-mg frozen liver samples from offspring were ground in liquid N2 and suspended in 50 ml of PBS. Protein-DNA cross-linking was performed by 37 % formaldehyde in 50 ml of PBS buffer for 10 min at room temperature. The cross-linking reaction was stopped with 2 m glycine. The following steps are the same as described by Strakovsky et al. (Reference Strakovsky, Wang and Engeseth21). Purified DNA was quantified by real-time PCR (the primers are listed in Table 2).
Statistical analysis
Results are presented as the mean values with their standard error of mean. Food intake and body weight were analysed by repeated-measures ANOVA. mRNA levels, protein levels and chromatin immunoprecipitation data were analysed using two-way ANOVA to look at the effects of maternal diets, sex and their interactions on the hepatic responses in the offspring, specially changes in the mechanisms that regulate autophagy-related gene expression (F-C and F-LP, M-C and M-LP, sample size n 8 for each). When a significant interaction was observed, a Bonferroni post hoc test was conducted to determine the effect of maternal diet on female (F-C v. F-LP) and male (M-C v. M-LP) offspring separately. All analyses were conducted in Prism 7. Differences were considered significant at P < 0·05 for all comparisons.
The sample size was calculated based on AAR gene expression data from a previous study(Reference Wang, Wilson and Zhou16) to achieve 80 % statistical power and 0·05 two-sided significance level.
Result
Physiological changes and growth potentials
Pregnant dams were obtained on day 2 of gestation. Fig. 2(a) shows dams’ daily food intake from gestation day 3 to day 21, at which they gave birth. There were no differences in the daily food intake and body weight (Fig. 2(b)) at the end of gestation between dams in the control and low protein groups. However, dams’ liver weights in the low protein group were slightly but significantly lower than those in the control group (Fig. 2(c)). At birth, pups in the control group weighed similar to pups in the LP group. As the pups grew to 21 d, the average body weight of female pups in the LP group was significantly lower than those in the control group (Fig. 2(d)). The average weight of male offspring in the LP group was not significantly different from the control pups (Fig. 2(e)). Liver weight and food intake did not differ between the control and LP pups (not shown).
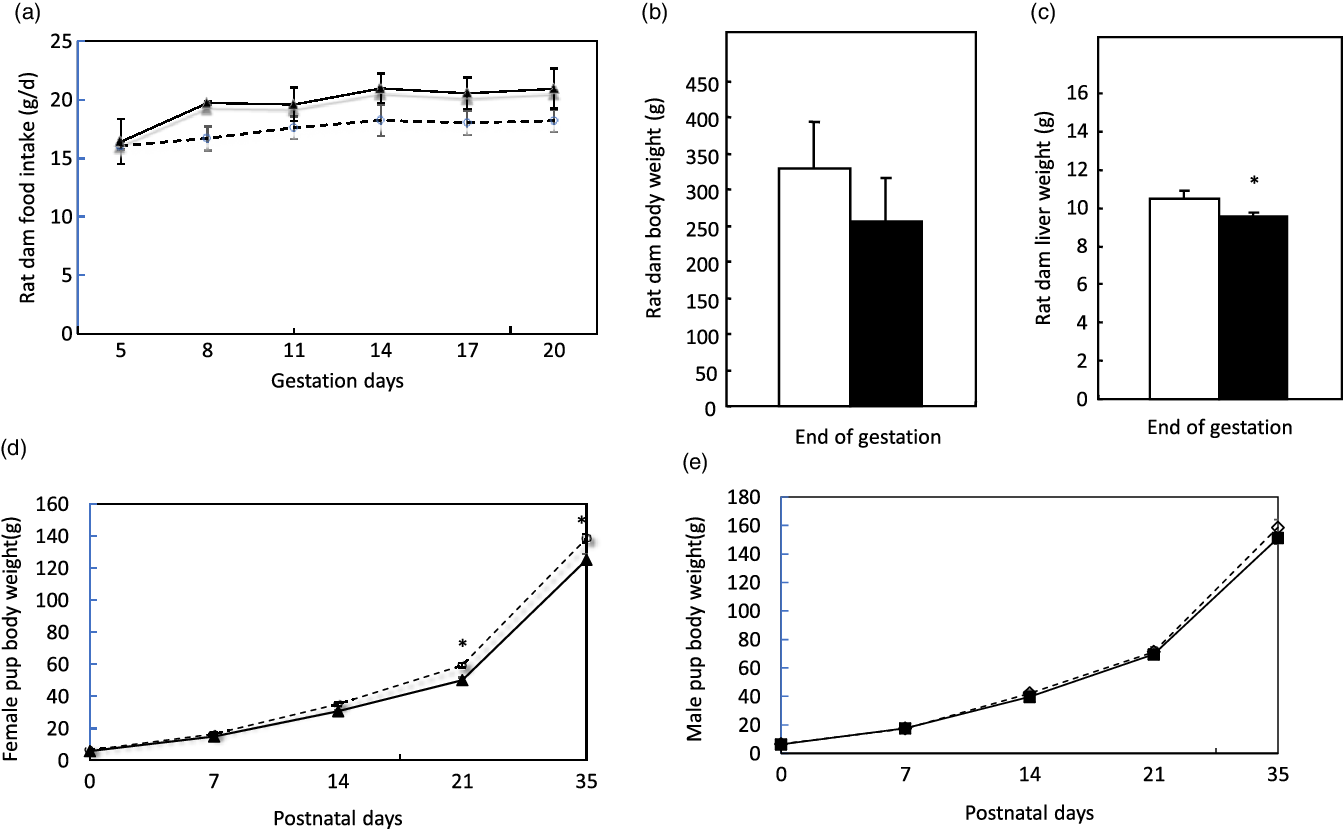
Fig. 2. Food intake (a), body weight (b), liver weight (c) of pregnant dams and body weight of female (d) and male pups (e) in the study. Values are reported as the mean values with their standard error of mean, n 8. *P < 0·05 analysed by repeated-measures ANOVA. C, pregnant dams fed on a control diet; LP, pregnant dams fed on a low-protein diet. (a) ![]() , C;
, C; ![]() , LP. (b)
, LP. (b) ![]() , C;
, C; ![]() , LP. (c)
, LP. (c) ![]() , C;
, C; ![]() , LP. (d)
, LP. (d) ![]() , C;
, C; ![]() , LP. (e)
, LP. (e) ![]() , C;
, C; ![]() , LP.
, LP.
Expression of amino acid response pathway-related genes
Two-way ANOVA demonstrated a significant interaction between maternal diet and offspring sex. Post hoc analysis showed that two markers of the AAR pathway, Atf3 (P = 0·003) and Chop (P < 0·001), exhibited significantly greater expression in the F-LP pups than in the F-C pups. However, there was no significant difference between the M-C and the M-LP pups (Fig. 3).
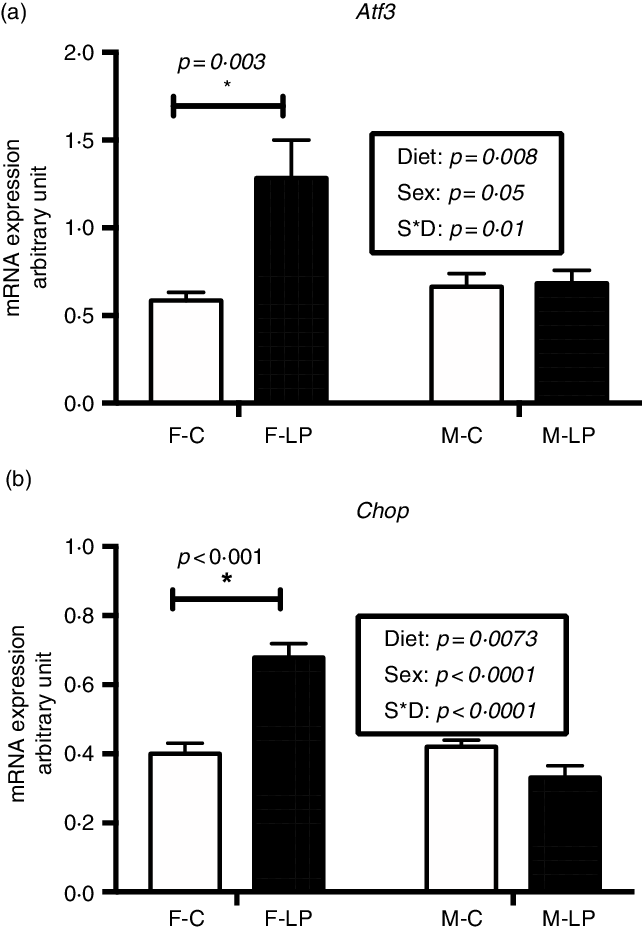
Fig. 3. A maternal LP diet induces the mRNA expression of Atf3 (a) and Chop (b) only in the liver of female offspring. All data were normalised to the expression level of the housekeeping genes L7a. Values are reported as the mean values with their standard error of mean, n 8. *P < 0·05 by post hoc Bonferroni test. Atf3, activating transcription factor 3; Chop, gene encoding C/EBP homology protein; L7a, gene encoding 60S ribosomal protein. F-C, female offspring born to the dams of control diet; F-LP, female offspring born to the dams of low-protein diet; M-C, male offspring born to the dams of control diet; M-LP, male offspring born to the dams of low-protein diet. (a) ![]() , C;
, C; ![]() , LP. (b)
, LP. (b) ![]() , C;
, C; ![]() , LP.
, LP.
Activation of the eukaryotic translation initiation factor 2/activating transcription factor 4 pathway
The eIF2a/ATF4 pathway was activated in the livers from the F-LP pups (Fig. 4). Two-way ANOVA showed a significant interaction between maternal diet and offspring sex. Therefore, a post hoc analysis was conducted and indicated that the protein levels of p-eIF2α (P = 0·002) and ATF4 (P < 0·001), which regulate the transcription of autophagy-related genes, were greater in the livers from the F-LP offspring than in those from the F-C offspring. The pathway was not activated in the livers from M-C and M-LP offspring (Fig. 4).
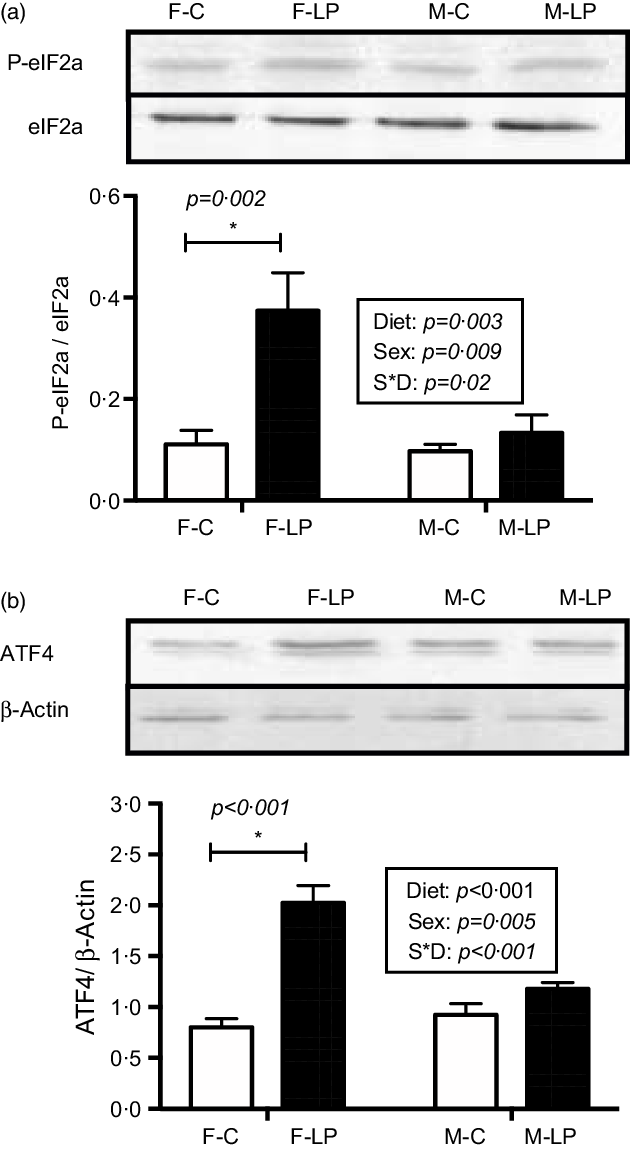
Fig. 4. The maternal LP diet activates the protein levels of p-eIF2a (a) and ATF4 (b) in the female offspring. Total eIF2a was used to normalise the phosphorylation of eIF2a, and actin was used to normalise the protein levels of ATF4. Values are reported as the mean values with their standard error of mean, n 8. *P < 0·05 by post hoc Bonferroni test. p-eIF2a, phosphorylated eukaryotic translation initiation factor 2a; ATF4, activating transcription factor 4. F-C, female offspring born to the dams of control diet; F-LP, female offspring born to the dams of low-protein diet; M-C, male offspring born to the dams of control diet; M-LP, male offspring born to the dams of low-protein diet. (a) ![]() , C;
, C; ![]() , LP. (b)
, LP. (b) ![]() , C;
, C; ![]() , LP.
, LP.
Expression of autophagy-related gene expression
The female pups born to LP dams had increased mRNA levels of hepatic autophagy-related genes than those born to C dams (Fig. 5). There was a substantial interaction effect between maternal diet and offspring sex on the mRNA expression levels of a set of autophagy-related genes. In female offspring, the mRNA expression of Becn1 (P = 0·04), Atg3 (P < 0·001), Map1lc3b (P = 0·001), P62/Sqstm1 (P < 0·001), Atg7 (P = 0·02), Atg5 (P < 0·001) and Atg10 (P < 0·001) was significantly higher in the F-LP group than in the F-C group. In male offspring, there was no effect of maternal diet observed (Fig. 5).
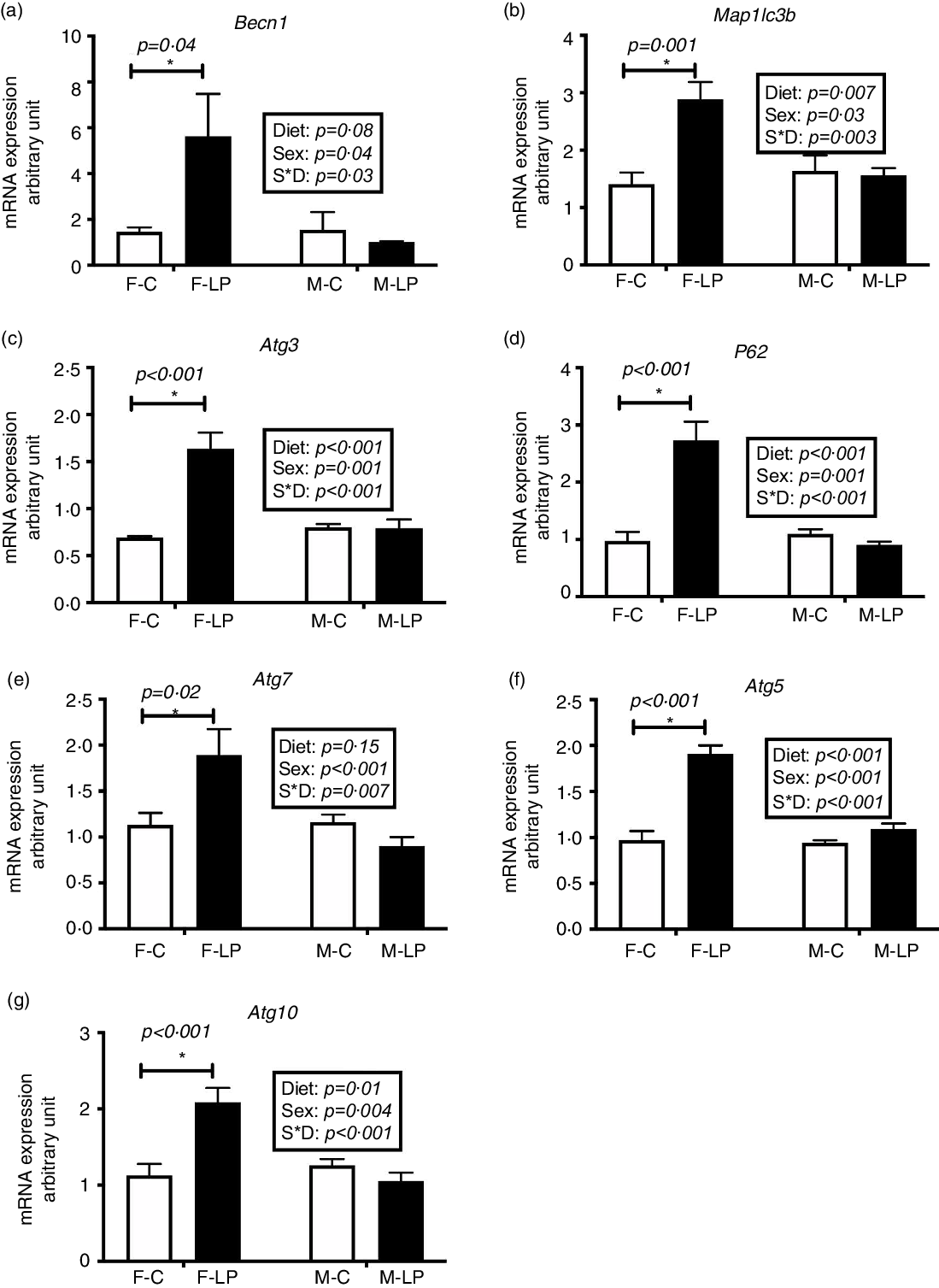
Fig. 5. The mRNA expression of hepatic autophagy-related genes including Becn1 (a), Map1lc3b (b), Atg3 (c), P62 (d), Atg7 (e), Atg5 (f) and Atg10 (g) was greater significantly in the female rat offspring of dams fed a maternal LP diet than those of dams fed a control diet. All data were normalised to the mRNA expression level of L7a. Values are reported as the mean values with their standard error of mean, n 8. *P < 0·05 by post hoc Bonferroni test. L7a, gene encoding 60S ribosomal protein. F-C, female offspring born to the dams of control diet; F-LP, female offspring born to the dams of low-protein diet; M-C, male offspring born to the dams of control diet; M-LP, male offspring born to the dams of low-protein diet. (a) ![]() , C;
, C; ![]() , LP. (b)
, LP. (b) ![]() , C;
, C; ![]() , LP. (c)
, LP. (c) ![]() , C;
, C; ![]() , LP. (d)
, LP. (d) ![]() , C;
, C; ![]() , LP. (e)
, LP. (e) ![]() , C;
, C; ![]() , LP. (f)
, LP. (f) ![]() , C;
, C; ![]() , LP. (g)
, LP. (g) ![]() , C;
, C; ![]() , LP.
, LP.
The role of activating transcription factor 4-C/EBP homology protein in autophagy-related gene expression
A post hoc test was conducted in female and male offspring separately due to a significant interaction between maternal diet and offspring sex. ATF4 and CHOP cooperatively regulated the transcriptional induction of autophagy-related genes in the livers in the female offspring. From the chromatin immunoprecipitation results, we found that: (1) ATF4 but not CHOP bound at the regions of Becn1 (P < 0·001), Atg3 (P = 0·01) and Map1lc3b (P < 0·001) (Fig. 6); (2) both ATF4 and CHOP bound to P62/Sqstm1 (both P < 0·001) and Atg7 (both P < 0·001) (Fig. 7); and (3) CHOP bound in the regions of Atg5 (P < 0·001) and Atg10 (P < 0·001) (Fig. 8). However, none of these bindings was observed differently in any male offspring (Figs. 6–8).
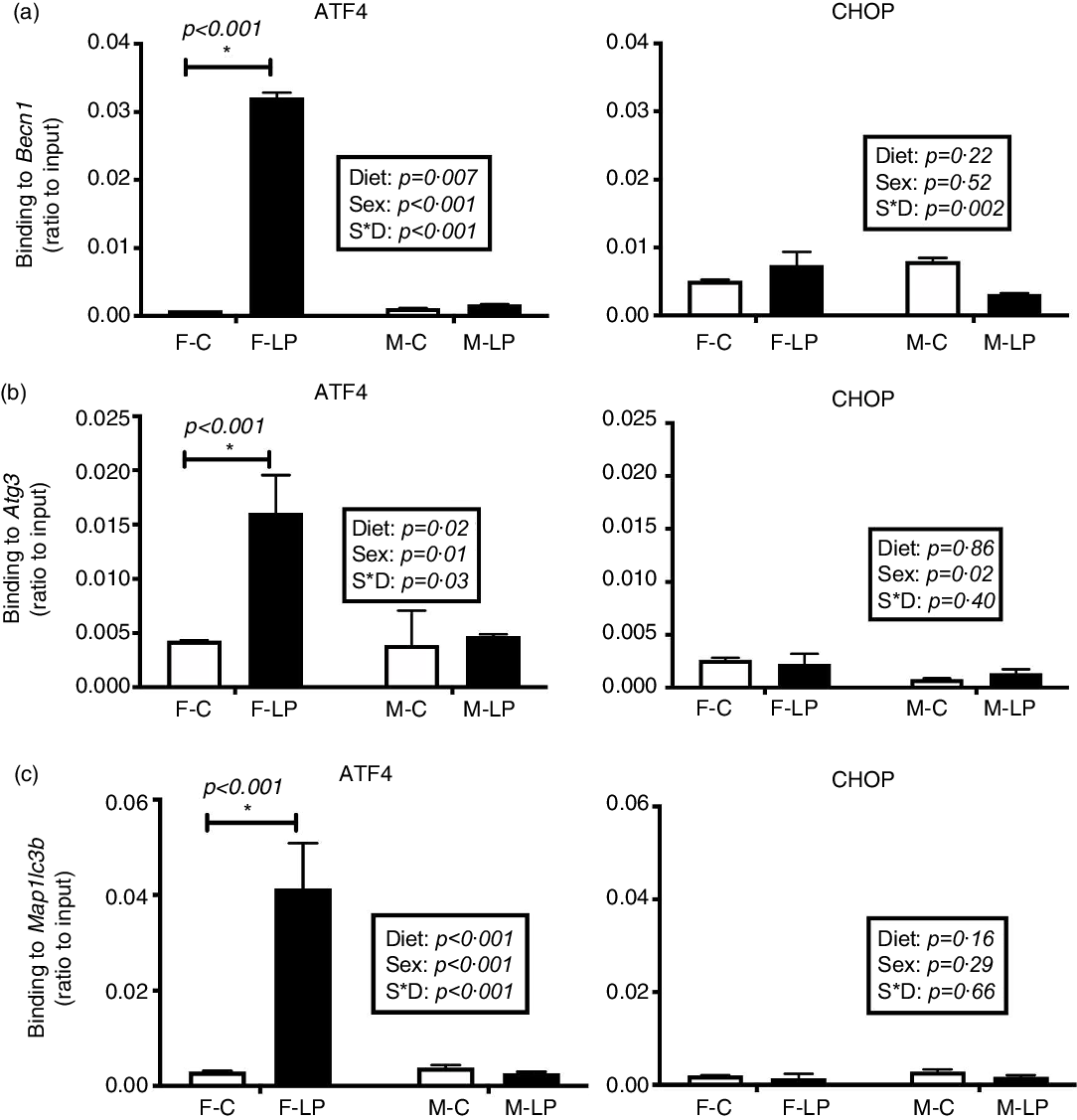
Fig. 6. The transcription factor ATF4 but not CHOP bound to Becn1 (a), Atg3 (b) and Map1lc3b (c) in the liver of female offspring of dams fed a maternal LP diet. Values are reported as the mean values with their standard error of mean, n 8. *P < 0·05 by post hoc Bonferroni test. ATF4, activating transcription factor 4; CHOP, C/EBP homology protein. F-C, female offspring born to the dams of control diet; F-LP, female offspring born to the dams of low-protein diet; M-C, male offspring born to the dams of control diet; M-LP, male offspring born to the dams of low-protein diet. (a) ![]() , C;
, C; ![]() , LP. (b)
, LP. (b) ![]() , C;
, C; ![]() , LP. (c)
, LP. (c) ![]() , C;
, C; ![]() , LP.
, LP.

Fig. 7. The binding of both ATF4 and CHOP to the autophagy-related genes P62 (a) and Atg7 (b) was induced in the liver of female offspring of dams fed a maternal LP diet. Values are reported as the mean values with their standard error of mean, n 8. *P < 0·05 by post hoc Bonferroni test. ATF4, activating transcription factor 4; CHOP, C/EBP homology protein. F-C, female offspring born to the dams of control diet; F-LP, female offspring born to the dams of low-protein diet; M-C, male offspring born to the dams of control diet; M-LP, male offspring born to the dams of low-protein diet. (a) ![]() , C;
, C; ![]() , LP. (b)
, LP. (b) ![]() , C;
, C; ![]() , LP.
, LP.
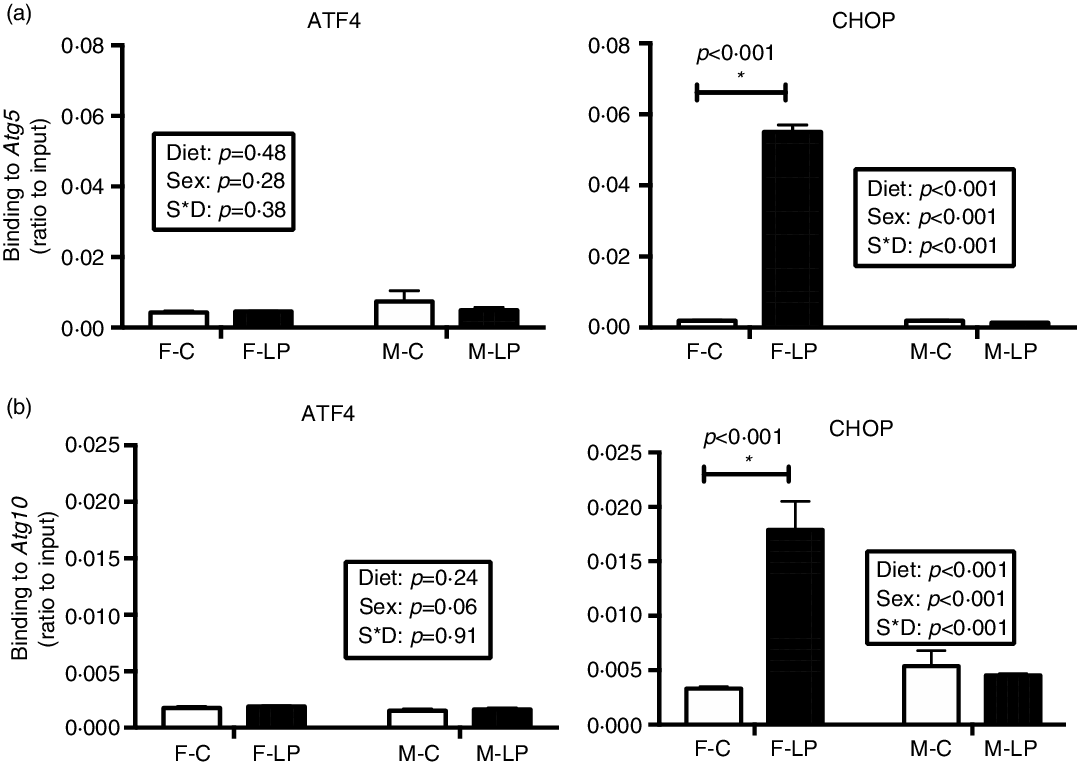
Fig. 8. The maternal LP diet induced the transcription factor CHOP but not ATF4 to bind to Atg5 (a) and Atg10 (b) in the liver of female offspring. Values are reported as the mean values with their standard error of mean, n 8. *P < 0·05 by post hoc Bonferroni test. ATF4, activating transcription factor 4; CHOP, C/EBP homology protein. F-C, female offspring born to the dams of control diet; F-LP, female offspring born to the dams of low-protein diet; M-C, male offspring born to the dams of control diet; M-LP, male offspring born to the dams of low-protein diet. (a) ![]() , C;
, C; ![]() , LP. (b)
, LP. (b) ![]() , C;
, C; ![]() , LP.
, LP.
Increased protein levels in the autophagy initiation
The protein levels of some of the genes in the autophagy pathway were detected in the female offspring exposed to a maternal LP diet (Fig. 9). A post hoc test was conducted within each sex due to a significant interaction between maternal diet and offspring sex. In female offspring, protein levels of Beclin 1 (P < 0·001) and LC3-II (P < 0·001), which are the primary indicators of the initiation in the autophagy pathway, were significantly higher in the F-LP group than in the F-C group. In male offspring, the maternal diet had no significant effect on these two protein levels (Fig. 9).

Fig. 9. The protein levels of Beclin1 (a) and LC3-II (b) were induced by the maternal LP diet only in the liver of female offspring. All data were normalised to the protein levels of Actin. Values are reported as the mean values with their standard error of mean, n 5. *P < 0·05 by post hoc Bonferroni test. LC3, microtubule-associated protein 1 light chain 3. F-C, female offspring born to the dams of control diet; F-LP, female offspring born to the dams of low-protein diet; M-C, male offspring born to the dams of control diet; M-LP, male offspring born to the dams of a low-protein diet. (a) ![]() , C;
, C; ![]() , LP. (b)
, LP. (b) ![]() , C;
, C; ![]() , LP.
, LP.
Discussion
Using an in vivo LP model, the present study demonstrates that maternal protein restriction during gestation affects the expression of autophagy-related genes in the liver of female offspring only. In response to a gestational LP diet, eIF2α was phosphorylated, which further activated ATF4. The activated eIF2α/ATF4 pathway induced autophagy-related gene expression through cooperative regulation of ATF4 and CHOP. A set of autophagy-related genes could be identified and divided into three classes according to their dependence on ATF4 and CHOP. These were only observed in the livers from female offspring but not from the male. This study provides the first direct evidence suggesting that maternal protein restriction during gestation induces hepatic autophagy-related gene expression in a sex-specific manner in the offspring. The induction of autophagy-related gene expression may result in an increased capacity to undergo autophagy in the liver of female offspring (Fig. 10).
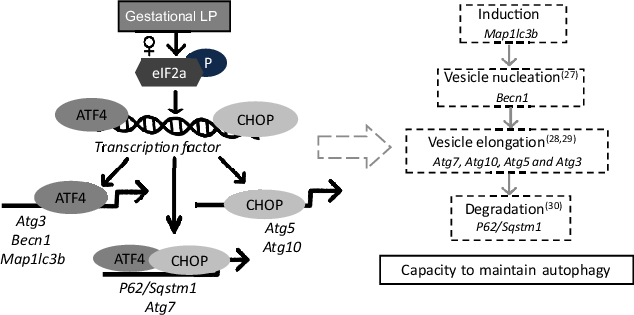
Fig. 10. A schematic model of autophagy-related gene transcriptional programming by maternal protein restriction in the liver of female rat offspring. It was adapted and redrawn from B’Chir et al. (Reference B’Chir, Maurin and Carraro11) with permission. In response to a low-protein diet during gestation, eIF2a was phosphorylated in the liver of female offspring, which further induced the transcription factors ATF4 and CHOP. ATF4 and CHOP then established a transcriptional programme of autophagy-related gene expression. The activated autophagy-related genes might result in an increased capacity to maintain autophagy by encoding autophagy-related proteins that can directly participate in the autophagic process. ATF4, activating transcription factor 4; CHOP, C/EBP homology protein.
Autophagy has been widely reported recently in different physiological conditions. However, understanding of the mechanisms that control autophagy pathways is limited, especially at the transcriptional level. This study focused on two major regulators, ATF4 and CHOP, which activate the transcription of the target genes Atg5 and Map1lc3b during hypoxia(Reference Rouschop, van den Beucken and Dubois22) and ER stress(Reference Pike, Singleton and Buffa23,Reference Raciti, Lotti and Valia24) . Nevertheless, there is limited research into their transcriptional roles in autophagy induced by nutrient stress. Furthermore, the downstream target autophagy-related genes have not been fully investigated. Recently, B’Chir et al. proposed a mechanism underlying autophagy-related gene expression regulated by eIF2α/ATF4 in mouse embryonic fibroblast cells upon amino acid starvation(Reference B’Chir, Maurin and Carraro11). This study suggested that ATF4 and CHOP were required to induce the transcription of a set of autophagy-related genes. Three classes of autophagy-related genes were clustered according to their different dependences on ATF4 and CHOP(Reference B’Chir, Maurin and Carraro11). Based on the autophagy transcriptional programme found by B’Chir, the present study further investigated the transcriptional roles of ATF4 and CHOP in a rat offspring model in which dams were fed on an LP diet during gestation. In the present study, cooperative regulation by ATF4 and CHOP resulted in three types of transcriptional programmes: (i) ATF4 but not CHOP bound to the promoter regions of Map1lc3b, Atg3 and Becn1 and up-regulated the transcription of these genes; (ii) both ATF4 and CHOP were observed to bind at the regions of P62/Sqstm1 and Atg7, which further activated their gene expression; and (iii) CHOP binding at the corresponding promoter induced the gene expression of Atg5 and Atg10 without direct interaction with ATF4. This investigation of the ATF4/CHOP-regulated programme will improve the understanding of the transcriptional regulation of autophagy pathways.
Autophagy pathways involve induction, nucleation, formation, elongation and degradation of autophagosomes(Reference Huang and Klionsky25,Reference Kraft and Martens26,Reference Kang, Zeh and Lotze27,Reference Nakatogawa28,Reference Shpilka, Mizushima and Elazar29,Reference Pankiv, Clausen and Lamark30) . The increased protein levels of Beclin 1 and LC3-II, as well as the elevated mRNA expression of a set of autophagy-related genes in a sex-specific effect in our study, may suggest an increased capacity to undergo autophagy in the liver of female offspring (Fig. 10). However, the cause is not fully understood. This difference potentially originated from the placenta through sex-dependent epigenetic regulation(Reference Dearden, Bouret and Ozanne31). Many diseases show alterations in autophagy and sex-associated differences(Reference Lista, Straface and Brunelleschi32,Reference Vitale, Mendelsohn and Rosano33,Reference Vega-Naredo, Caballero and Sierra34) . Different degrees of autophagy depending on sex may be controlled by the redox-sensitive transcription factors, such as NF-κB and p53 in an organ-specific way(Reference Vega-Naredo, Caballero and Sierra34,Reference Campesi, Straface and Occhioni35) . Several hepatic genes could respond to maternal protein restriction in a sex-specific manner, including G6PC (Reference Jia, Cong and Li36), PEPCK (Reference Kwong, Miller and Wilkins37) and 11beta-HSD1 (Reference Kwong, Miller and Wilkins37). Hormonal regulation may also contribute to sex dimorphism, including oestrogen played an essential role in autophagy regulation in the liver and exhibited female-biased response(Reference Mohapatra, Chakraborty and Shimizu38,Reference Xiang, Liu and Ren39) . Moreover, maternal protein restriction could impair androgen levels in male offspring(Reference Toledo, Perobelli and Pedrosa40,Reference Zambrano, Rodriguez-Gonzalez and Guzman41) and influenced liver metabolism(Reference Shen and Shi42). The different features displayed in this study could be mainly due to hormonal regulators, as previously suggested(Reference Maselli, Matarrese and Straface43). However, how hormonal regulators influence ATF4 and CHOP binding to specific genes is largely unknown at this moment. It is also possible that timing for the postpartum analysis may be important. For example, males may show a similar response, but this response occurs earlier or later (before/after current postpartum analysis). More research must be conducted to investigate the role of epigenetics and sex hormones in offspring to understand how maternal nutrition differentially impacts males and females fully.
The sex-specific responses in hepatic autophagy-related genes may result in differential physiological outcomes and potential diseases. Autophagy-related genes have been linked to human cancers and other diseases. For instance, Becn1 has been reported to be a tumour-suppressor gene. In a Becn1-mutant mouse model, the development of hepatitis B virus-induced premalignant lesions was significantly accelerated(Reference Qu, Yu and Bhagat44). Another example is the role of Atg7 in lipid metabolism in the liver of mice. In Atg7-knockout mice, a massive accumulation of lipid droplets has been found in the liver, suggesting that without Atg7, mice cannot undergo autophagy to degrade unnecessary lipids and have an elevated risk of non-alcoholic fatty liver disease(Reference Singh, Kaushik and Wang45). Several sex-biased diseases can be explained to some extent by sex-specific autophagy-related gene expression. For example, differences in sex-specific autophagy-related gene expression have been found in cardiac samples from both mice and humans, potentially explaining the disparity in heart disease between the sexes found in clinical studies(Reference Koenig, Sateriale and Budd46). Our findings highlight the sex difference in autophagy-related gene expression in rat offspring of dams exposed to gestational protein restriction, suggesting that male and female offspring might have different hepatic conditions. Future studies should investigate the hepatic physiological outcomes in male and female offspring experiencing maternal protein restriction, especially the potential sex disparity in metabolic phenotype. Furthermore, the lack of activated autophagy-related gene expression in male offspring suggests that male offspring potentially have a reduced ability to adapt to a poor maternal environment(Reference Cheong, Wlodek and Moritz47,Reference Bromfield, Schjenken and Chin48,Reference Isganaitis, Woo and Ma49) . As a result, males might have a greater risk of adverse outcomes(Reference Isganaitis, Woo and Ma49,Reference Gawlinska, Gawlinski and Korostynski50,Reference Spearman, Ke and Fu51) and need more clinical attention than females. Understanding the sex disparity in physiological effects in offspring may help diagnose potential sex-biased diseases that occur in response to a maternal LP diet during pregnancy.
Acknowledgements
The present study was supported by funding from the UIUC Research Board (grant no. RB14148) and the Division of Nutritional Sciences (Vision 20/20 grant) and supported by the USDA Cooperative State Research, Education and Extension Service (project no. #ILLU-698-374). However, the sponsors had no role in the design and analysis of the study or in the writing of this article.
The authors’ responsibilities were as follows – M. Z. C. and Y. X. P.: Conceptualization; M. Z. C. and J. Z.: Data curation; M. Z. C.: Formal Analysis; H. C.: Funding acquisition, Resources; M. Z. C. and J. Z.: Writing – original draft; M. Z. C. and Y. X. P.: Writing – review & editing and all authors read and approved the final manuscript. The final manuscript has been proofread by the American Journal Experts.
The authors who have taken part in this study declare that they have no conflicts of interest concerning this manuscript.
















