During pregnancy, maternal macronutrient metabolism changes to adapt to the needs of the developing fetus( Reference Rieger, Loskutoff and Betteridge 1 ). Although changes in glucose metabolism occur in parallel with increasing energy demands, adaptation of protein metabolism – through the accumulation and breakdown of protein stores – is an essential component of maternal adaptation to pregnancy( Reference Kalhan 2 ). Pregnancy is often associated with hypoaminoacidaemia and urinary N losses during fasting, which is evident early in gestation and persists throughout pregnancy( Reference Metzger, Unger and Freinkel 3 , Reference Felig, Kim and Lynch 4 ). The major source of urea N during pregnancy is peripheral (muscle) release of amino acids( Reference Kalhan 2 ), and Kalkhoff et al. ( Reference Kalkhoff, Kandaraki and Morrow 5 ) showed a positive correlation between total circulating amino acid concentrations and neonatal birth weight. The role of maternal protein balance in both maternal health and gestational outcomes highlights the importance of the maternal diet. Because of this, the low-protein (LP) rodent model is one of the best-characterised developmental programming models, demonstrating that even a marginal protein restriction (8 % of energy from protein) during pregnancy has physiological impacts on both the mother and the offspring( Reference Snoeck, Remacle and Reusens 6 ).
Dietary protein restriction in experimental animals and cell-culture models produces numerous metabolic responses, including the activation of the amino acid response (AAR) pathway( Reference Kilberg, Shan and Su 7 ). AAR pathway activation in response to amino acid limitation occurs largely through the signal transduction-mediated altered transcriptional activity of specific genes including Asns (asparagine synthetase)( Reference Barbosa-Tessmann, Chen and Zhong 8 ), Atf3 (activating transcription factor 3)( Reference Pan, Chen and Siu 9 ) and Chop (C/EBP homology protein)( Reference Abcouwer, Schwarz and Meguid 10 , Reference Bruhat, Jousse and Wang 11 ). The activation of the AAR pathway requires the phosphorylation of eukaryotic translation initiation factor 2α (eIF2α) by general control non-derepressible 2 (GCN2) kinase, leading to a decrease in global protein synthesis( Reference Lu, Harding and Ron 12 ) but an increase in the synthesis of activating transcription factor 4 (ATF4) protein( Reference Lu, Harding and Ron 12 – Reference Vattem and Wek 14 ), which activates multiple stress-induced genes, including genes containing an AAR element( Reference Harding, Zhang and Zeng 15 ). Therefore, ATF4 appears to serve as a master regulator of the AAR pathway, further supported by observations that ATF4 binds at the promoter regions of Asns ( Reference Chen, Pan and Dudenhausen 16 ) and Atf3 ( Reference Pan, Chen and Thiaville 17 ).
Due to the well-characterised and direct relationship between protein restriction (even marginally) and the activation of ATF4 target genes, the AAR pathway has served as a hallmark marker of dietary protein deficiency. However, skeletal muscle accounts for over 40 % of body protein and is a critical site for labile protein in response to nutritional stress, and the autophagic/lysosomal pathway has also been associated with protein mobilisation in skeletal muscle( Reference Sandri, Lin and Handschin 18 ). Therefore, autophagy could potentially serve as an additional nutrient-sensing mechanism in response to protein deficiency. Autophagy, primarily known as macroautophagy in cellular metabolism, involves cell degradation of unnecessary or dysfunctional cellular components through the lysosomal machinery, and has been highly associated with myopathy( Reference Nishino 19 , Reference Mizushima, Levine and Cuervo 20 ). LC3 and BNIP3 have been shown to be key proteins during the process of autophagy( Reference Mammucari, Milan and Romanello 21 ), and ATF4, the key regulatory protein within the AAR pathway that has a role in muscle dystrophy induced by fasting( Reference Ebert, Dyle and Kunkel 22 , Reference Ebert, Monteys and Fox 23 ), has also recently been associated with autophagy( Reference Ebert, Dyle and Kunkel 22 , Reference B'Chir, Maurin and Carraro 24 ).
Gestational protein restriction activates the placental AAR pathway, which is associated with long-term growth restriction in the offspring( Reference Strakovsky, Zhou and Pan 25 ). However, the response and potential contribution of maternal metabolism in this process is unknown. Therefore, the purpose of the present study was to investigate the molecular response of maternal tissues to gestational protein restriction. Specifically, we first focused on the AAR pathway as the classic marker of protein restriction, and then investigated the potential role of ATF4 within the autophagy pathway, as a potential physiological consequence of gestational protein deficiency. We also relied on our study in which gestationally LP diet-exposed offspring were growth-restricted adults, despite consuming a control diet after weaning, to determine whether gestational protein restriction was still evident at the molecular level in the muscle of these offspring at postnatal day 38. Understanding the adaptations that occur within maternal metabolism in response to protein restriction would provide valuable insight into our understanding of how the maternal tissue/placental/fetal axis contributes to the programming of chronic diseases in response to maternal protein deficiency.
Experimental methods
Animals and diets
Timed-pregnant Sprague–Dawley rats were obtained from Charles River Laboratories and randomly assigned to one (n 6) of two isoenergetic, modified AIN-93 diets( Reference Reeves, Nielsen and Fahey 26 ) (Research Diet, Inc.; Table 1). Study 1 included the following diets: control diet (20 % of energy from protein, 64 % of energy from carbohydrate and 16 % of energy from fat) or low-protein diet (LP; 8 % of energy from protein, 76 % of energy from carbohydrate and 16 % of energy from fat) from day 2 of gestation until the end of gestation. Study 2 included the following diets: control diet (18 % of energy from protein, 66 % of energy from carbohydrate and 16 % of energy from fat) or LP diet (9 % of energy from protein, 75 % of energy from carbohydrate and 16 % of energy from fat) from day 2 of gestation until the end of gestation( Reference Strakovsky, Zhou and Pan 25 ). Rat dams in study 2 consumed a control diet during lactation( Reference Strakovsky, Zhou and Pan 25 , Reference Zheng and Pan 27 ), and all offspring consumed the same control diet after weaning (postnatal day 21) until postnatal day 38. The rats were housed individually in standard polycarbonate cages in a humidity- and temperature-controlled environment at a 12 h light–12 h dark cycle. Body weights and food intake were recorded every other day.
Table 1 Composition of the experimental diets*
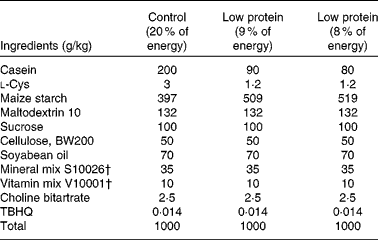
TBHQ, tertiary butylhydroquinone.
* Modified AIN-93 G diet.
† Vitamin mix V10001 and mineral mix S10026 were obtained from Dyets, Inc.
Post-delivery dams were fasted for 8 h and then euthanised by CO2 for 20 s before decapitation to collect blood for plasma and serum preparation. Visceral adipose tissue, gastrocnemius muscle and liver samples from dams in study 1 were harvested and snap-frozen in liquid N2 and stored at − 80°C for future analyses. Skeletal muscle from the offspring was collected on 38 d after birth in study 2. All experiments were conducted in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals( Reference Bayne 28 ), and approved by the University of Illinois at Urbana-Champaign Institutional Animal Care and Use Committee.
Plasma measurements
Maternal plasma obtained from trunk blood was immediately centrifuged at 800 g for 20 min at 4°C. Plasma samples were stored at − 70°C until further analysis. Plasma free amino acid concentrations were measured by HPLC using a Waters 2475 fluorescence detector (Waters)( Reference Reverter, Lundh and Lindberg 29 ). Amino acid hydrolysate standard was purchased from the same company (WAT088122). Analyses included essential amino acids (Leu, Ile, Lys, Met, Val and Thr) and non-essential amino acids (His, Glu, Asp, Pro, Tyr, Ala, Arg, Ser and Gly).
Muscle fibre diameter
From each treatment group, six frozen muscle samples were embedded in Tissue-Tek OCT compound (VWR) and cut into 5 μm thickness cross-sectionally using a Leica CM3050 S cryostat (Leica Microsystems, Inc.) at − 20°C, as reported previously( Reference Zhang, Zhou and Strakovsky 30 , Reference Kintscher, Hartge and Hess 31 ). All sections were fixed in 70 % ethanol and stained with haematoxylin and eosin (Newcomer Supply). Histopathological examination was performed blindly using light microscopy by a certified pathologist (S. L.). Slides were scanned using a NanoZoomer Digital Pathology System with a magnification of 20 × (Hamamatsu Photonics K.K.), and image analysis was performed using ImageJ software. From each section, three independent regions were selected for quantification. The diameters of at least sixty myofibres per independent region were measured using the lesser diameter method, as described previously( Reference Ebert, Monteys and Fox 23 ).
RNA isolation and two-step real-time quantitative PCR
Total RNA was isolated from the visceral adipose tissue, gastrocnemius muscle, liver and blood of dams, and from the gastrocnemius muscle of the offspring using TRI-Reagent (Sigma-Aldrich), according to the manufacturer's instructions. RNA concentration was analysed by a Bio-Rad spectrophotometer (Bio-Rad Laboratories, Inc.) at 260 nm. Total mRNA (2 μg) was used for complementary DNA synthesis by the High Capacity cDNA Reverse transcription Kit (Applied Biosystems) with a DNA 2720 Thermal Cycler (Applied Biosystems). The programme was as follows: 25°C for 10 min, 37°C for 120 min and 85°C for 5 s. After synthesis, 25 ng of complementary DNA were analysed using real-time quantitative PCR, and gene expression was assessed using the 2 × Perfecta SYBR Green Fast Master Mix (Quanta BioSciences, www.vwr.com) with a 7300 Real-Time PCR System (Applied Biosystems). Primer sequences used in the experiment are listed in online Supplementary Table S1, and were designed by Vector NTI software (Invitrogen Corporation) and synthesised by Integrated DNA Technologies (www.idtdna.com). The reaction was as follows: 95°C for 15 min, 95°C for 15 s and 60°C for 60 s in forty cycles. After amplification, dissociation curves were generated by stepwise increases from 55 to 95°C to ensure that a specific product was amplified in the reaction. Standard curves with a slope of − 3·30 (sem 0·20) and R 2≥ 0·99 were accepted. The following tissue-specific housekeeping genes whose expression was not affected by the treatment were used to normalise all mRNA data: 60S ribosomal protein (L7a) in rat dams' adipose tissue and in offspring skeletal muscle; TATA-binding protein (Tbp) in rat dams' liver and blood; β-actin (Actb) in rat dams' skeletal muscle.
Protein isolation and Western blotting
Frozen gastrocnemius muscle (200 mg) was ground in liquid N2 and suspended in 500 μl of protein lysis buffer (0·125 m-Tris–HCl, pH 6·8, 1 % SDS, 0·04 % bromophenol blue and 20 % glycerol, v/v) with 1 × proteinase inhibitor (Roche Applied Science) and phosphatase inhibitor cocktail 1 and 2 (Sigma-Aldrich). Samples were then sonicated with twenty-five pulses at a power setting of 2 (Fisher Scientific Model 100 Sonic Dismembrator). Diluted protein samples (20 μg) were size-fractionated on a 12 % Tris–HCl polyacrylamide gel and transferred onto a polyvinyl difluoride (PVDF) membrane (Bio-Rad Laboratories, Inc.) at 0·3 A, according to the wet transfer protocol. To investigate ATF4 (CAMP response element-binding protein 2; CREB2) protein expression, the PVDF membrane was incubated with blocking solution (10 % (w/v) non-fat dry milk (NFDM)) in TBS/T (20 mm-Tris–HCl, pH 7·6, 137 mm-NaCl and 0·1 % (v/v) Tween-20) for 1 h at room temperature. A rabbit polyclonal antibody against ATF4 (sc-200; Santa Cruz) was diluted at 1:1500 in the blocking solution with 10 % NFDM and incubated with the membrane for 3 h at room temperature before washing (5 × ) with 5 % NFDM in TBS/T for 5 min. A goat anti-rabbit HRP-conjugated secondary antibody (Kirkegaard&Perry Laboratories) was diluted at 1:10 000 in the blocking solution containing 5 % NFDM and incubated with the membrane for 1 h at room temperature. After washing for 5 min in the blocking solution (5 × ) containing 3 % (w/v) NFDM, the membrane was exposed to the enhanced chemiluminescence reagent Super Signal West Dura (Thermo Fisher Scientific). Signals from the membrane were detected and quantified by the ChemiDoc XRS Imaging System (Bio-Rad). β-Actin (SC-1616-R; Santa Cruz) was used as a loading control for the protein expression of ATF4. Total eIF2α (9722S; Cell Signaling Technology, Inc.) was used as a loading control for phosphorylated eukaryotic translation initiation factor 2α (p-eIF2α) (9721L; Cell Signaling Technology, Inc.). The condition for detecting the p-eIF2α protein was as follows: 5 % NFDM in TBS/T solution for 1 h of blocking and 1:1000 diluted primary antibody against p-eIF2α for 3 h of incubation at room temperature. A blocking buffer of 1 % NFDM in TBS/T solution was used for washing (5 × ) for 5 min followed by incubation with a secondary monoclonal goat-anti-rabbit antibody (Cell Signaling Technology, Inc.) for 1 h. Furthermore, 1 % NFDM in TBS/T solution was used for the subsequent 5 min washing (5 × ).
Chromatin immunoprecipitation
Chromatin immunoprecipitation analysis was performed according to a modified protocol( Reference Chen, Pan and Dudenhausen 16 ). In brief, 200 mg frozen gastrocnemius muscle was ground in liquid N2 and suspended in PBS. Protein–DNA cross-linking was performed by 1 % formaldehyde in PBS solution for 10 min, and the reaction was stopped by 2 m-glycine to a final concentration of 0·125 m. Nuclei were collected after nuclei swelling buffer treatment (5 mm-piperazine-N,N′-bis, 2-ethanesulfonic acid (PIPES), pH 8·0, 85 mm-KCl, 0·5 % NP40) and lysed in SDS lysis buffer (50 mm-Tris–HCl, pH 8·1, 10 mm-EDTA, 1 % SDS) containing protease inhibitor and phosphatase inhibitor cocktails. Cross-linked chromatin was sheered and homogenised on ice by a Sonic Dismembrator (Model F100; Fisher Scientific) for 40 s at a power setting of 5 with a 2 min cooling interval between each burst, with a total of five bursts. The average length of sonicated chromatin was approximately 500 bp, and cell debris was removed by centrifugation at 13 000 g for 10 min at 4°C. Sheared chromatin was diluted to 10 ml with chromatin immunoprecipitation dilution buffer. A solution of 1 ml diluted chromatin was incubated overnight with 2 μg antibody at 4°C. The chromatin–antibody complex was precipitated with 60 μl of 50 % slurry of pre-blocked protein G-agarose beads (Millipore) for 2 h at 4°C. A normal rabbit IgG was used as a negative control. The supernatant from the incubated IgG was used as the input DNA for each sample. The protein G-agarose beads were sequentially washed with 1 ml each of the following solutions: low salt (20 mm-Tris–HCl, pH 8·0, 0·1 % SDS, 2 mm-EDTA, 150 mm-NaCl, 1 % Triton X-100), high salt (20 mm-Tris–HCl, pH 8·0, 0·1 % SDS, 2 mm-EDTA, 500 mm-NaCl, 1 % Triton X-100), LiCl (10 mm-Tris–HCl, pH 8·0, 0·25 mm-LiCl, 1 % NP40, 1 mm-EDTA, 1 % sodium deoxycholate); twice with Tris–EDTA (pH 8·0). The antibody/protein/DNA complexes were eluted twice by the addition of 250 μl elution buffer (1 % SDS and 50 mm-NaHCO3) at 37°C for 15 min each time. Reverse cross-linking was performed with 20 μl of 5 m-NaCl and 1 μg of RNase A at 65°C for 5 h. Chromatin DNA was purified using the QIAPrep Miniprep Kit (Qiagen) after proteinase K digestion. Results are expressed as the average ratio of IgG to input DNA. The antibodies are listed in online Supplementary Table S2.
Bioinformatics
Transcription factor binding sites were identified by TFSEARCH( Reference Heinemeyer, Wingender and Reuter 32 ) and MatInspector( Reference Cartharius, Frech and Grote 33 ) programs. All sequences were derived from the promoter sequence retrieval database EIDorado 02-2010 (Genomatix). The motif of the AAR element and CAMP response element-binding protein (CREB) was scanned with the Gene2Promoter large-scale program in the Genomatix software suite GEMS Launcher version 2.7 to evaluate any genes containing AAR element and CREB binding motifs within the rat genome( Reference Cartharius, Frech and Grote 33 ). The promoter sequences were defined as those predicted in EIDorado and elongated at the 3′ end of the promoter (downstream) by 150 bp. Position weight and matrices were used according to the Matrix Family Library version 8.2 (Genomatix, January 2010) for promoter analysis (Fig. 4(a)).
Statistical analysis
Data are presented as means and standard deviations. Food intake and body weight were analysed by repeated-measures one-way ANOVA (SAS Institute, Inc.). Two-way ANOVA was used for chromatin immunoprecipitation data with a Tukey post hoc comparison. Student's t test was used for two-group comparisons (control v. LP), including plasma amino acid concentrations, mRNA expression and protein expression. Differences were considered significant at P< 0·05 and P< 0·01. Sample size was calculated by statistical power analysis based on 95 % CI using Statistical Solutions, LLC.
Results
Gestational low-protein diet reduces weight gain in rat dams
Pregnant rats consuming a LP diet weighed significantly less than the control group at days 17, 19 and 21 of gestation (P< 0·01; Fig. 1(a)), but food intake was not significantly different in LP rat dams compared with the control group (Fig. 1(b)). Gestational weight gain was significantly lower in the LP group than in the control group (P< 0·01, Fig. 1(c)). Litter sizes were not different between the control and LP groups. The birth weight of LP offspring was significantly lower (P< 0·05, Fig. 1(d)) than that of the control group. However, even after taking offspring birth weight into consideration, net gestational weight gain (after subtracting pup birth weight) remained significantly lower in LP dams when compared with the control group (P< 0·01, Fig. 1(e)).

Fig. 1 Body weight (a), food intake (b), gestational weight gain (c), offspring birth weight (d) and gestational net weight gain (e) of rat dams consuming a control (–Δ–) or low-protein (LP, –■–) diet throughout gestation in study 1. Values are means (n 6), with their standard errors represented by vertical bars. Where not visible, the error bars are too small to be seen. Mean value was significantly different from that of the control group: * P< 0·05, ** P< 0·01.
Gestational low-protein diet alters the plasma amino acid profile in rat dams
At the end of pregnancy, circulating Thr and His levels were significantly lower (P< 0·05) in LP dams than in the control group (Table 2). Lys, Ala and Ser were significantly higher (P< 0·05) in the LP group, while Leu, Ile, Lys, Met, Val, Glu, Asp, Pro, Tyr, Arg and Gly did not differ between the groups (see online Supplementary Table S3).
Table 2 Plasma amino acid profile of rat dams in study 1 (Mean values with their standard errors; n 6)
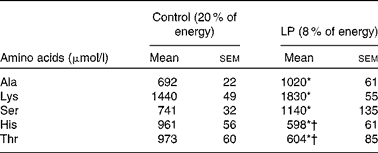
* Mean value was significantly different from that of the control group (P< 0·05).
† Indicates decreased values.
Gestational low-protein diet induces skeletal muscle atrophy in rat dams
Muscle consists of bundles of multinucleated cells, called muscle fibres or myofibres, which were assessed in rat dams after delivery using haematoxylin and eosin. A gestational LP diet has been reported to alter the mechanical properties of skeletal muscle in male offspring( Reference Toscano, Manhaes-de-Castro and Canon 34 ); however, the consequences for the dam have not been reported. After delivery, we observed more varied sizes of muscle fibres, splitting muscle fibres and clustered basophilic fibres in the skeletal muscle of the LP group when compared with the control group (Fig. 2(a)). Fibres in the size range of 46–65 μm were decreased in the LP group when compared with the control group (P< 0·05), whereas the LP group had more fibres in the size range of 66–91 μm when than the control group (Fig. 2(b)). Overall, the gestational LP diet caused a more varied distribution of fibre diameters than the control diet (Fig. 2(c)) in the skeletal muscle of dams. Furthermore, the gestational LP diet significantly decreased the overall average skeletal muscle fibre size in rat dams (Fig. 2(d)), which has previously been associated with myopathy( Reference Ebert, Dyle and Kunkel 22 , Reference Ebert, Monteys and Fox 23 , Reference Fox, Ebert and Bongers 35 ).

Fig. 2 Haematoxylin and eosin (H&E) staining (a), quantification (b, d) and distribution analysis (c) of the skeletal muscle fibres of rat dams consuming a control or low-protein (LP) diet throughout gestation in study 1. H&E staining was used to investigate the structural changes in the skeletal muscle of rat dams after a gestational LP diet. Two representative images were chosen to show the structural differences in skeletal muscle between the control and LP groups at a magnification of 20 × . ![]() , Clustered basophilic fibres;
, Clustered basophilic fibres; ![]() , varied sizes of muscle fibres and splitting muscle fibres. For quantification of muscle fibre percentage by different diameter ranges, each bar comprises the percentage distribution of fibre number in four different ranges of muscle fibre diameters, including 0–45 (□), 46–65 (
, varied sizes of muscle fibres and splitting muscle fibres. For quantification of muscle fibre percentage by different diameter ranges, each bar comprises the percentage distribution of fibre number in four different ranges of muscle fibre diameters, including 0–45 (□), 46–65 (![]() ), 66–91 (
), 66–91 (![]() ) and 92–140 μm (■). The X-axis represents the treatment group, while the Y-axis shows the percentage of fibre number in each diameter range. Values are means (n 6), with their standard errors represented by vertical bars. * Mean value was significantly different from that of the control group (P< 0·05). (c) –Δ–, Control group; –■–, LP group. The X-axis shows a series of fibre diameters and the Y-axis shows the percentage of fibre number. (d)
) and 92–140 μm (■). The X-axis represents the treatment group, while the Y-axis shows the percentage of fibre number in each diameter range. Values are means (n 6), with their standard errors represented by vertical bars. * Mean value was significantly different from that of the control group (P< 0·05). (c) –Δ–, Control group; –■–, LP group. The X-axis shows a series of fibre diameters and the Y-axis shows the percentage of fibre number. (d) ![]() , Control group; ●, LP group. (A colour version of this figure can be found online at http://www.journals.cambridge.org/bjn).
, Control group; ●, LP group. (A colour version of this figure can be found online at http://www.journals.cambridge.org/bjn).
Gestational low-protein diet induces amino acid response pathway-related target genes in the skeletal muscle of rat dams
The impact of gestational protein restriction on the AAR pathway was assessed by investigating the mRNA expression of AAR downstream genes (Asns, Atf3 and Chop) in the visceral adipose tissue, gastrocnemius (skeletal) muscle, liver and blood of rat dams after delivery. The gestational LP diet induced the mRNA expression of Asns (2-fold, P< 0·05), Atf3 (1-fold, P< 0·05) and Chop (1·5-fold, P< 0·05) when compared with the control diet in skeletal muscle (Fig. 3(a)) but not in other tissues (liver Atf3 expression: P= 0·08 for the LP diet; Fig. 3(b)). These results show that the gestational LP diet induces a tissue-specific response of AAR downstream genes in rat dams.

Fig. 3 mRNA expression of Asns (asparagine synthetase), Atf3 (activating transcription factor 3) and Chop (C/EBP homology protein) in the muscle (a), liver (b), adipose (b) and blood (b) of rat dams consuming a control (□) or low-protein (LP, ■) diet throughout gestation in study 1. To detect the activation of the amino acid response (AAR) pathway, mRNA expression was analysed by two-step real-time RT-PCR in skeletal muscle. All data were normalised to the expression level of the housekeeping gene β-actin (Actb) (muscle) or 60S ribosomal protein (L7a) (liver, adipose and blood), and data from the LP group were further normalised to that of the control group. Values are means (n 6), with their standard errors represented by vertical bars. Mean value was significantly different from that of the control group: * P< 0·05.
Gestational low-protein diet increases activating transcription factor 4 protein expression and phosphorylation of eukaryotic translation initiation factor 2α as well as activating transcription factor 4 binding at the promoter regions of amino acid response genes in the skeletal muscle of rat dams
To confirm the activation of the AAR pathway, the protein abundance of whole-cell p-eIF2α and ATF4 was analysed in the skeletal muscle of dams after delivery (Fig. 4(a) and (b)). Phosphorylation of eIF2α on Ser 51, an upstream event required for the activation of the AAR pathway, was higher in the skeletal muscle of the LP group compared with the control group (Fig. 4(a) and (b)), and muscle ATF4 protein expression was significantly higher in the LP group than in the control group (Fig. 4(a) and (b)). Based on the observed increased mRNA expression of ATF4 target genes, we investigated the binding of ATF4 at the promoter regions of these genes in the skeletal muscle of dams after delivery. Overall, ATF4 binding was significantly induced by the gestational LP diet at the predicted regions of the genes examined, including Asns (3-fold, P< 0·05), Atf3 (1·5-fold, P= 0·06) and Chop (4·5-fold, P< 0·05) (Fig. 4(c)), thus confirming the ATF4-induced activation of the AAR pathway in the muscle of rat dams at the end of gestation.

Fig. 4 Protein expression of phosphorylated eukaryotic translation initiation factor 2α (p-eIF2α) (a, b), activating transcription factor 4 (ATF4) (c, d), and ATF4 binding at the promoter regions of Asns (asparagine synthetase), Atf3 (activating transcription factor 3) and Chop (C/EBP homology protein) (d) in rat dams consuming a control (□) or low-protein (LP, ■) diet throughout gestation in study 1. Total eIF2α was used to normalise the phosphorylation of eIF2α, and β-actin was used to normalise the protein expression of ATF4. The immunoblots shown are representative of the total sample size. Values are means (n 6), with their standard errors represented by vertical bars. Mean value was significantly different from that of the control group: * P< 0·05, ** P< 0·01. AAR, amino acid response.
Gestational low-protein diet increases the mRNA expression of autophagy pathway-related genes and LC3B protein expression in the skeletal muscle of rat dams
The proposed process of cellular autophagy and its relationship with the AAR pathway is described in Fig. 5(a) (adapted from the literature( Reference Mammucari, Milan and Romanello 21 , Reference B'Chir, Maurin and Carraro 24 , Reference Masiero, Agatea and Mammucari 36 )). The mRNA expression of autophagy-related genes, including LC3a (1·5-fold, P< 0·05,), LC3b (2-fold, P< 0·05), Bnip3 (2·5-fold, P< 0·05) and Atg4b (1·4-fold, P< 0·05), was significantly induced in dams fed the gestational LP diet (Fig. 5(b)). Furthermore, to confirm the observed activation of autophagy, the protein abundance of whole-cell LC3B was analysed in the skeletal muscle of dams. The protein expression of LC3B, which was used as the primary indicator of autophagy, was higher in the skeletal muscle of LP dams when compared with the control group (2-fold, P< 0·05; Fig. 5(c)).

Fig. 5 Schematic diagram of cellular autophagy (a), autophagy gene expression (b) and LC3B protein expression (c) in the skeletal muscle of rat dams consuming a control (□) or low-protein (LP, ■) diet throughout gestation in study 1. To detect the activation of the autophagy pathway, mRNA expression was analysed by two-step real-time RT-PCR in skeletal muscle. All data were normalised to the expression level of the housekeeping gene β-actin (Actb), and data from the LP group were further normalised to that of the control group. β-Actin was used to normalise the protein expression of LC3B. The immunoblots shown are representative of the total sample size. Values are means (n 6), with their standard errors represented by vertical bars. Mean value was significantly different from that of the control group: * P< 0·05. p-eIF2α, phophorylated eukaryotic translation initiation factor 2α; ATF4, activating transcription factor 4; Asns, asparagine synthetase; Chop, C/EBP homology protein; LC3a, microtubule-associated protein 1 light chain 3α; LC3b, microtubule-associated protein 1 light chain 3β; Bnip3, BCL2/adenovirus E1B-interacting protein 3; Atg4b, cysteine protease ATG4b.
Gestational low-protein diet induces activating transcription factor 4 binding at the regions of autophagy pathway-related genes predicted by bioinformatics analysis in the skeletal muscle of rat dams
Based on the increased mRNA expression of autophagy-related genes and the proposed relationship between autophagy and AAR, we investigated ATF4 binding at the predicted regions of downstream autophagy pathway-related genes in the skeletal muscle of dams (Fig. 6(a)). When compared with the control, the gestational LP diet in dams increased ATF4 binding at the predicted regions of the autophagy-related genes examined, including LC3a (2-fold, P< 0·01), LC3b (2-fold, P< 0·05), Atg4b (2·5-fold, P< 0·01) and Bnip3 (1·5-fold, P< 0·05) (Fig. 6(b)).

Fig. 6 Binding of activating transcription factor 4 (ATF4) to autophagy-related genes (a, b) in rat dams consuming a control (□) or low-protein (LP, ■) diet throughout gestation in study 1. The binding sites of the ATF4 transcription factor at the promoter regions of LC3a (microtubule-associated protein 1 light chain 3α), LC3b (microtubule-associated protein 1 light chain 3β), Atg4b (cysteine protease ATG4b) and Bnip3 (BCL2/adenovirus E1B-interacting protein 3) were predicted by Genomatix software. Values are means (n 6), with their standard errors represented by vertical bars. Mean value was significantly different from that of the control group: * P< 0·05, ** P< 0·01.
Gestational low-protein diet induces amino acid response pathway- and autophagy pathway-related genes in the skeletal muscle of male offspring
Our previous study showed that the placenta may sense gestational protein limitation, which is associated with growth restriction in the offspring( Reference Strakovsky, Zhou and Pan 25 ). To investigate whether these previous observations and the observation of the present study that maternal skeletal muscle is potentially involved in response to gestational protein limitation, we investigated the mRNA expression of both AAR- and autophagy-related genes in the skeletal muscle of offspring in our previously described study. Similar to what was observed in dams at delivery, the gestational LP diet significantly induced the mRNA expression of Asns (2-fold, P< 0·01), Atf3 (5-fold, P< 0·05) and Chop (2-fold, P< 0·01) in male, but not in female, offspring skeletal muscle (Fig. 7(a)). Furthermore, and also similar to that observed in the dam, the mRNA expression of autophagy-related genes, including LC3a (3-fold, P< 0·05), LC3b (5-fold, P< 0·01), Atg2a (2-fold, P< 0·05) and Atg4b (4-fold, P< 0·05), was significantly induced by the gestational LP diet in male, but not in female, offspring skeletal muscle (Fig. 7(b)).

Fig. 7 mRNA expression of amino acid response (AAR) pathway-related genes (a) and autophagy pathway-related genes (b) in the skeletal muscle of offspring whose dams consumed a control (□) or low-protein (LP, ■) diet throughout gestation in study 2. To detect the activation of the AAR pathway and autophagy in the offspring, mRNA expression was analysed by two-step real-time RT-PCR in skeletal muscle. All data were normalised to the expression level of the housekeeping gene 60S ribosomal protein (L7a), and data from the maternal LP group were further normalised to that of the control group. Values are means (n 6), with their standard errors represented by vertical bars. Mean value was significantly different from that of the control group: * P< 0·05, ** P< 0·01. Asns, asparagine synthetase; Atf3, activating transcription factor 3; Chop, C/EBP homology protein; Atg2a, cysteine protease ATG2a; Atg4b, cysteine protease ATG4b.
Discussion
To our knowledge, the present study is the first to demonstrate the following novel mechanisms: first, a gestational LP diet alters the maternal plasma amino acid profile, together with decreased maternal and pup body weights and apparent muscle dystrophy in the dam at delivery in an in vivo rat pregnancy model. Second, the LP diet produced tissue-specific activation of AAR and autophagy pathway-related genes only in maternal skeletal muscle, but not in the liver, blood or adipose tissue. Third, the LP diet increased ATF4 binding within specific autophagy-related target genes, confirming a molecular link between the activation of the AAR signal and the autophagy pathway. Together, these data, as well as the follow-up data showing that in the skeletal muscle of male offspring fed a gestation LP diet, AAR- and autophagy-related genes remain elevated, provide insight into the transduction of the gestational amino acid limitation signal from maternal muscle, to the placenta( Reference Strakovsky, Zhou and Pan 25 ), and to the offspring. A proposed schematic diagram highlights the mechanism that a gestational LP diet programmed offspring growth capacity through signal transduction from the skeletal muscle of the rat dam (Fig. 8). Furthermore, the molecular mechanisms described in the present study offer novel insight into the role of ATF4 in activating critical autophagy-related genes in the skeletal muscle of pregnant dams, providing specific molecular changes that could be memorised in the offspring long after gestational protein restriction.

Fig. 8 Diagram of the proposed maternal programming mechanism by a gestational low-protein diet in rat dams. The results highlight that a gestational low-protein diet activates the amino acid response (AAR) pathway and potentially initiates autophagy in the skeletal muscle of the rat dam. The signal is potentially transduced to the placenta and further to the skeletal muscle of male offspring, which may potentially link to the stunted growth in the offspring. Asns, asparagine synthetase; Atf3, activating transcription factor 3; Snat2, sodium-coupled neutral amino acid transporter 2; Chop, C/EBP homology protein; LC3a/b, microtubule-associated protein 1A/1B light chain 3α/β; Bnip3, BCL2/adenovirus E1B-interacting protein 3; Atg4b, cysteine protease ATG4b; AA, amino acids.
The present study demonstrates that consumption of a LP diet throughout pregnancy altered multiple physiological outcomes in rat dams. Gestational weight gain was decreased in LP dams (after accounting for cumulative offspring birth weight), as were the birth weight of the LP offspring. It has been reported that exposure of the pregnant rat to a LP diet (8 % of energy from protein) causes a decrease in maternal body weight without a significant difference in food intake( Reference Chamson-Reig, Thyssen and Hill 37 ), which is consistent with the findings from the present study. Fasting blood glucose levels of LP rat dams (3·53 mmol/l) tended to be lower (P= 0·06) than the control group (6·28 mmol/l; data not shown), which is in agreement with the findings of Wapnir & Lifshitz( Reference Wapnir and Lifshitz 38 ) showing that a LP diet can cause a decrease in fasted blood glucose concentration in rats. Furthermore, muscle fibre perimeter was overall significantly different in LP dams when compared with the control group. Previous studies have demonstrated that decreased fibre size is linked to myopathy( Reference Fox, Ebert and Bongers 35 ), and our findings suggest that gestationally protein-restricted dams adapt to the growing protein needs of pregnancy through the breakdown of maternal muscle.
We observed that a gestational LP diet was associated with significant decreases in maternal plasma Thr concentration, consistent with the findings from a previous study showing that protein limitation decreases circulating Thr concentration in pregnant rats( Reference Rees, Hay and Buchan 39 ). There were also changes in Lys, Ala and Ser concentrations in the LP group, consistent with other studies reporting increased mobilisation of maternal protein stores during dietary restriction( Reference Wu, Pond and Ott 40 ). The increased plasma concentration of the essential amino acid Lys cannot be explained by de novo synthesis, and can only result from increased protein breakdown or reduced hepatic catabolism. The breakdown of maternal protein stores by a gestational LP diet is further confirmed by the observed increase in Ala concentration, which has a central role in gluconeogenesis and serves to remove N and recycle glucose carbon as part of the ‘alanine cycle’ associated with branched-chain amino acid oxidation in skeletal muscle( Reference Felig 41 ). Increased Ala and Lys concentrations have also been previously observed in a pregnant pig model of protein restriction( Reference Felig 41 ).
At the end of gestation, the LP diet induced the activation of AAR and autophagy pathways in maternal skeletal muscle. The mechanism of AAR pathway activation in response to amino acid deprivation has been well documented in in vitro studies( Reference Kilberg, Shan and Su 7 , Reference Kilberg, Pan and Chen 42 ), but remains poorly characterised in animals. Decreased Thr concentration has been previously shown to activate the AAR pathway in HepG2 cells( Reference Palii, Kays and Deval 43 ). In another study, a Thr-deficient diet has been found to activate the AAR pathway in an animal model( Reference Hao, Ross-Inta and Gietzen 44 , Reference Hao, Sharp and Ross-Inta 45 ). The present study demonstrates that decreases in plasma Thr and His concentrations are primary indicators of protein limitation in maternal plasma after gestation, consistent with in vitro data for the activation of the AAR pathway( Reference Palii, Kays and Deval 43 ). Our results show that a robust activation of the AAR pathway occurred only in skeletal muscle following a gestational LP diet, which was illustrated by the increased mRNA expression of the AAR-responsive genes and ATF4 and p-eIF2α protein expression. Previous research has shown that increased ATF4 expression promoted skeletal myofibre atrophy during fasting( Reference Ebert, Monteys and Fox 23 ). Moreover, Naismith & Morgan( Reference Naismith and Morgan 46 ) found that protein redistribution occurs during pregnancy, and it has also been shown that muscle has the greatest role during this period of active protein metabolism( Reference Naismith and Morgan 46 – Reference Naismith 48 ). While amino acid availability is associated with the regulation of autophagy in yeast and mammalian cells( Reference Abeliovich 49 ), in vivo data are scarce. In the present study, we showed that the LP diet induced the activation of the autophagy pathway in the skeletal muscle of rat dams. Macroautophagy is highly associated with cellular metabolism, and autophagy in skeletal muscle has both beneficial and detrimental roles, depending on the stage of protein metabolism( Reference Sandri 50 ). It has been reported that starvation can induce autophagy in the muscle of mice( Reference Mizushima, Yamamoto and Matsui 51 ). Furthermore, It has been shown that eIF2α/ATF4 plays an important role in activating stress-induced autophagy gene expression in vitro ( Reference B'Chir, Maurin and Carraro 24 ), and data from the present study demonstrate that autophagy is also activated in the skeletal muscle of pregnant rats consuming a protein-deficient diet.
In support of the ATF4-induced gene expression in muscle, the binding of the ATF4 transcription factor was consistently increased at the promoter regions of both AAR and autophagy pathway-related genes in response to the LP diet. We observed the binding of ATF4 at the promoter regions of Asns, Atf3 and Chop, which was associated with the induction of mRNA expression similar to the ATF4-mediated activation of the AAR pathway in vitro in hepatoma cells( Reference Kilberg, Shan and Su 7 ). Increased ATF4 binding also occurred at the promoter regions of LC3a, LC3b, Atg4b and Bnip3 along with increased mRNA expression of these autophagy pathway-related genes. ATF4 is known to regulate the hypoxia-inducible factor-1 (HIF-1) target gene carbonic anhydrase 9. Here, we show that another HIF-1 target gene, Bnip3 ( Reference van den Beucken, Koritzinsky and Niessen 52 , Reference Guo, Searfoss and Krolikowski 53 ), can also be regulated by ATF4. Furthermore, we observed increased LC3B protein expression, which has been used as a biomarker for the activation of autophagy in muscle( Reference Kazi and Lang 54 ). These data demonstrate that in addition to the role of ATF4 in the activation of the AAR pathway in response to protein restriction, this transcription factor also activates autophagy pathway-related genes in the muscle of protein-deficient pregnant dams. Furthermore, while gestational protein restriction has previously been shown to affect the skeletal muscle of offspring( Reference Mortensen, Olsen and Frandsen 55 , Reference Zhu, Ford and Nathanielsz 56 ), we further demonstrate that both AAR and autophagy pathways remain activated in male offspring long after the exposure to a gestational LP diet, suggesting that the induction of autophagy through the activation of the ATF4-dependent AAR pathway in the mother may act as the molecular memory of the forthcoming expected environment in the offspring. A LP diet during pregnancy could be memorised in the offspring in different ways. From physiological point of view, during pregnancy, the fetus obtains amino acid resource through the umbilical cord from the placenta. Therefore, an altered amino acid profile in mother has a direct influence on the amino acid profile in the fetus. It has been shown that amino acid profile was altered in the fetus on pregnancy day 19 by maternal LP diet( Reference Rees, Hay and Buchan 39 ). From molecular point of view, altered amino acid supply triggers subsequential signalling pathways in embryonic development. A LP diet during pregnancy can programme blastocyst development, which is potentially due to the activation of the mammalian target of rapamycin (mTOR) signalling pathway triggered by altered maternal serum amino acids in mice( Reference Eckert, Porter and Watkins 57 ). Moreover, epigenetics may be the key to explain the ‘memory’. The maternal programming model has been well established in a gestational LP diet to investigate epigenetic regulation including histone modification and DNA methylation. The effect of epigenetics is tissue- and sex-dependent. Histone acetylation of glucose transporter type 4 (Glut4) was induced by a gestational LP diet in the skeletal muscle of female offspring( Reference Zheng, Rollet and Pan 58 ), and the diet decreased histone H3 acetylation and methylation of cyclin-dependent kinase inhibitor 1A (p21) in the mammary gland of the offspring( Reference Zheng, Rollet and Yang 59 ). Also, increased hepatic DNA methylation of insulin-like growth factor 2 (Igf2) could be caused by a LP diet during pregnancy( Reference Gong, Pan and Chen 60 ). It appears that the effects of the maternal LP diet may be memorised in the offspring through multiple epigenetic mechanisms.
Sex dimorphism of fetal programming has been studied intensively over the years. It potentially originates from the placenta through sex-specific epigenetic regulations( Reference Gabory, Roseboom and Moore 61 ). Therefore, male and female respond differently to maternal protein restriction depending on the type of metabolic pathways and tissues. A LP diet during gestation can induce Glut4 expression in the skeletal muscle of female offspring( Reference Zheng, Rollet and Pan 58 ), but no effect in male offspring. However, AAR and autophagy pathway-related genes are only up-regulated in the skeletal muscle of male offspring, possibly involve hormonal regulation. The development of male skeletal muscle is related to male hormones( Reference Michel and Baulieu 62 ). Moreover, it has been shown that a protein restriction diet during pregnancy and lactation impairs male testosterone levels( Reference Zambrano, Rodriguez-Gonzalez and Guzman 63 ). This may explain the differentiated transcriptional regulation of AAR and autophagy pathways in the skeletal muscle of male and female offspring.
The present study demonstrates that maternal protein restriction results in decreased Thr and His concentrations in plasma together with the activation of the ATF4-dependent AAR pathway, which may initiate autophagy in the skeletal muscle of rat dams. We showed that the transcriptional activation of target genes was associated with the binding of the ATF4 transcription factor at the promoter regions of these genes in AAR and autophagy pathways. Furthermore, a follow-up analysis of gestational protein restriction in the offspring confirmed that both pathways remained activated in the skeletal muscle of the offspring long after birth. Imbalanced and inadequate protein intake in women during pregnancy is an epidemic problem in developing countries( Reference Hartriyanti, Suyoto and Muhammad 64 , Reference Lee, Talegawkar and Merialdi 65 ). Recently, a new cohort in Canada showed that the requirement for protein intake is actually higher than the current recommendations in healthy pregnant women during early and late gestation( Reference Stephens, Payne and Ball 66 ). However, few epidemiological studies have focused on the response to maternal protein restriction in the mother. Not only the offspring, but also the mother has detrimental effects in response to protein restriction, and the present study sheds light on maternal healthcare for both women and babies. Classifying these protein-sensing mechanisms in the mother may help to diagnose potential muscle-associated diseases in response to LP intake during pregnancy, as well as to further investigate the precise role of maternal signalling in the growth restriction observed in the offspring in response to maternal protein deficiency.
Supplementary material
To view supplementary material for this article, please visit http://dx.doi.org/10.1017/S0007114515002172
Acknowledgements
The authors thank Dr Rita Strakovsky from the University of Illinois at Urbana-Champaign for copyediting and technical advice.
The present study was supported by funding from the UIUC Research Board (grant no. RB14148) and the Division of Nutritional Sciences (Vision 20/20 grant), and supported by the USDA Cooperative State Research, Education and Extension Service (project no. #ILLU-698-374). The sponsors had no role in the design and analysis of the study or in the writing of this article.
The contributions of the authors are as follows: the experiments were performed in the laboratory of Y.-X. P. at the University of Illinois at Urbana-Champaign; Y.-X. P. and H. W. were responsible for the conception and design of the experiments; H. W. and G. J. W. performed the analysis of amino acids by HPLC in the laboratory of D. K. L. at the University of Illinois at Urbana-Champaign; H. W. conducted all the experiments and collected the data; D. Z., S. L. and X. C. provided technical advice and assistance on the histological results; H. W. analysed the data, performed the statistical analysis and drafted the manuscript; Y.-X. P. and H. W. had the primary responsibility for the final content. All authors read and approved the final manuscript.
None of the authors has any conflict of interest to declare.













