Intensive systems of ruminant production encourage the inclusion of large proportions of grains or easily fermentable carbohydrates into the diet to support high milk yields or rapid weight gain. Although these feeding practices may be helpful in enhancing cost-efficiency in the short term, they have considerable consequences for animal gastrointestinal health. The effect of a high-grain (HG) diet on rumen health such as rumen acidosis has received increasing attention( Reference Liu, Xu and Liu 1 – Reference Steele, Croom and Kahler 3 ). However, to date, the effects of HG feeding on microbial fermentation processes and intestinal homeostasis in the hindgut of ruminants have received relatively little research attention, despite their importance in animal health and performance( Reference Gressley, Hall and Armentano 4 ).
Caecum is one of the main fermentation sites in the hindgut of ruminants. Faichney( Reference Faichney 5 ) showed that caecal fermentation accounts for 12 % of total volatile fatty acid (VFA) production in sheep. Dixon & Nolan( Reference Dixon and Nolan 6 ) reported that caecum is the major region of fermentation and absorption of the components of digesta in the large intestine of sheep. Thus, caecal fermentation in goats may make a considerable contribution to total metabolisable energy. Some recent studies have demonstrated that a HG diet increases caecal fermentation, which is characterised by high concentrations of VFA in the caecal digesta( Reference Gressley, Hall and Armentano 4 , Reference Li, Khafipour and Krause 7 ). These findings indicate that HG feeding may result in an increase in energy supply from the hindgut. However, due to the differences in buffering capacity between the rumen and the hindgut, an increase in the concentrations of caecal VFA may make the caecum less capable of maintaining digesta pH and lead to an unhealthy caecal environment( Reference Metzler-Zebeli, Schmitz-Esser and Klevenhusen 8 ). This detrimental luminal environment may also be associated with dramatic alterations in microbial compositions during HG feeding. However, no information is currently available regarding the changes in caecal luminal and mucosal microbial compositions caused by HG feeding. Therefore, as a response to dietary changes, adaptive alterations in microbial compositions in the caecum should be studied comprehensively to prevent the overlooking of these unfavourable dietary effects on the health of ruminants during HG feeding.
The gastrointestinal epithelium is known to represent a physical as well as an immunological barrier. As the first line of innate immune response, the gastrointestinal epithelium has been found to play an important role in the maintenance and regulation of gastrointestinal health. Previous studies have revealed that a HG diet increases the acidity and lipopolysaccharide (LPS) concentration of ruminal fluid and causes dramatic alterations in rumen microbial compositions, which, in turn, cause ruminal epithelial damage and increase epithelial permeability( Reference Steele, AlZahal and Hook 9 – Reference Chin, Flynn and Fedwick 11 ). Events that occur in the rumen during HG feeding are mirrored in the large intestine. As has been mentioned earlier, an increase in intestinal carbohydrate fermentation caused by HG feeding may lead to increased concentrations of short VFA and a reduction of pH in the caecum( Reference Li, Khafipour and Krause 7 ). The increased acidity may result in damage to and sloughing of the epithelial cells in the large intestine( Reference Gressley, Hall and Armentano 4 ). In addition, the intestinal epithelium is composed of only a single layer of epithelial cells, whereas the rumen epithelium consists of a multilayer squamous epithelium with significant keratinisation( Reference Plaizier, Khafipour and Li 12 ). Thus, differences between the structures of the intestinal and ruminal epithelia are likely to make the intestinal epithelium more susceptible to damage than the rumen during HG feeding. Using an equine model, Crawford et al. ( Reference Crawford, Sepulveda and Elliott 13 ) demonstrated that excessive hindgut fermentation causes inflammation and laminitis, probably due to the systemic entry of amines, endotoxins or bacteria through a breach in the intestinal barrier. In ruminants, however, despite the advances in our knowledge on hindgut metabolism during HG feeding, there is a lack of evidence in the published literature that increased caecal fermentation during HG feeding may cause similar damage to the intestinal lining and contribute to local inflammatory responses. We hypothesised that a HG diet increases the concentrations of VFA in the caecum while adversely affecting the make-up of caecal microbiota, which leads to a detrimental caecal environment, resulting in caecal mucosal injury and local inflammation. Therefore, the primary objective of the present study was to investigate the effect of HG feeding on caecal bacterial composition, caecal fermentation, mucosal morphology, and inflammatory cytokine expression in the mucosal epithelium of goats. Additionally, we also evaluated the relationship between the alterations in caecal fermentation and caecal mucosal bacterial composition and the expression of inflammatory cytokines in the mucosal epithelium.
Materials and methods
The experimental design and procedures were approved by the Animal Care and Use Committee of Nanjing Agricultural University in compliance with the Regulations for the Administration of Affairs Concerning Experimental Animals (The State Science and Technology Commission of People's Republic of China, 1988).
Animals, diets and experimental design
The present study was part of a larger experiment investigating the effect of a HG diet on the gastrointestinal health of goats( Reference Liu, Xu and Liu 1 ). The main study tested the hypothesis that a diet with a high proportion of grains will affect the health of the rumen and caecum. The details of this study have been reported previously( Reference Liu, Xu and Liu 1 ). Therefore, in the present study, we mainly focused on the effect of HG feeding on caecal fermentation, caecal microbiota and caecal epithelial morphometry.
A total of ten rumen-cannulated, castrated male goats (Boer × Yangtze River Delta White) aged 2–3 years were used in the experiment. Before the start of the experiment, all the goats were fed ad libitum a pure hay diet for 5 weeks to ensure adaptation to a low-energy hay diet. The goats were randomly assigned to two groups based on whether they were fed a hay diet (hay; 81 % Leymus chinensis, 15 % lucerne, 101 g crude protein/kg DM, and 570 g neutral-detergent fibre/kg DM; n 5) or a HG diet (HG; 30 % L. chinensis, 45 % maize meal, 20 % wheat meal, 1·1 % soyabean meal, 101 g crude protein/kg DM, 252 g neutral-detergent fibre/kg DM, and 582 g starch/kg DM; n 5) and were placed in individual pens (1·2 × 1·2 m) with free access to water. The diets (750 g DM/animal per d) were offered in equal amounts at 08.30 and 16.30 hours daily for 7 weeks (Table S1, available online). The metabolic energy intake was slightly above the requirement for the maintenance of goats in the hay group (8·31 MJ/kg DM) and permitted a growth rate of 200 g/d in goats of the HG group (30 kg body weight and 11·31 MJ/kg DM), according to guidelines on the nutrient requirement of goats (NY/Y816-2004; Ministry of Agriculture of China, 2004; Table S1, available online).
Sample collection
The experimental period lasted 7 weeks, and then the ten goats were slaughtered for sampling. The abdominal cavity was opened by midline incision and the gastrointestinal tract was removed carefully. A representative sample of rumen digesta (at least 200 ml) was used to determine the pH value and was then strained through four layers of cheesecloth. A sample of the ruminal fluid was preserved in 25 % (w/v) metaphosphoric acid (5 ml ruminal fluid:1 ml metaphosphoric acid) and stored at − 20°C until VFA analysis. Whole-caecum digesta were collected and homogenised, and then the pH of caecal digesta samples was determined immediately, using an Accumet gel-filled, polymer body combination pH electrode (Fisher Scientific). The homogenised digesta were divided into four portions. The first portion of each of the caecal digesta samples (10 g wet weight) was mixed thoroughly with double the amount of distilled water. The mixtures were immediately centrifuged at 2000 g and the supernatants were stored at − 20°C until VFA and lactic acid analyses. The second portion (10 g wet weight) was mixed with an equal amount of endotoxin-free water and the mixtures were immediately centrifuged at 13 000 g for 40 min at 4°C. The supernatants were boiled for 5 min and then stored at − 20°C until LPS analysis. The third portion (15 g wet weight) was dried at 60°C for 48 h. Dried samples were subsequently ground using a Wiley mill through a 1 mm screen and stored at − 20°C until starch analysis. The last portion was stored at − 80°C until DNA extraction.
Within 5 min of slaughter, a segment of the caecum was collected and immediately washed three times with ice-cold PBS. The segment was divided into three portions. The first portion was cut into smaller pieces of approximately 0·5 × 0·5 cm and the pieces were immediately frozen in liquid N2 for RNA extraction. The second portion was cut into pieces of approximately 2 × 2 cm and the pieces were scraped from the underlying tissue using a germ-free glass slide and immediately transferred into liquid N2 and then stored at − 80°C until DNA extraction. The last portion was immediately fixed in 4 % paraformaldehyde (Sigma) and 2·5 % glutaraldehyde for histomorphometric microscopy.
Physiological parameter measurements
The pH of the rumen liquid and caecal digesta was measured using a portable pH meter (HI 9125; HANNA Instruments), and the concentrations of VFA were measured using capillary column GC (GC-14B, Shimadzu; film thickness of the capillary column, 30 m × 0·32 mm × 0·25 mm; column temperature, 110°C; injector temperature, 180°C; and detector temperature, 180°C)( Reference Qin 14 ). The concentration of lactic acid was determined according to the method described by Barker & Summerson( Reference Barker and Summerson 15 ). The concentration of free LPS in the caecal digesta was measured using the Chromogenic End-point Tachypleus Amebocyte Lysate Assay Kit (Xiamen Horseshoe Crab Reagent Manufactory) with a minimum detection limit of 0·01 endotoxin unit/ml. Pretreated supernatants were diluted until their LPS concentrations were in the range of 0·1–1 endotoxin unit/ml relative to the reference endotoxin. The concentration of starch in the caecal digesta was determined using the Megazyme Total Starch Assay Kit (Megazyme International Ireland Limited).
Microbial DNA isolation
DNA was extracted using 1 g of wet caecal mucosa. DNA extraction was carried out using the bead-beating method with a mini-bead beater (Biospec Products), followed by phenol–chloroform extraction( Reference Mao, Zhang and Zhu 16 ). The solution was precipitated with ethanol, and the pellets were suspended in a 50 μl-Tris–EDTA buffer. The DNA samples were quantified using a NanoDrop spectrophotometer (Nyxor Biotech).
DNA pyrosequencing
Universal 16S ribosomal RNA (rRNA) gene primers (Escherichia coli positions 8–533: E8F 5′-AGAGTTTGATCCTGGCTCAG-3′ and E533R 5′-TTACCGCGGCTGCTGGCAC-3′) were chosen for the amplification and subsequent pyrosequencing of PCR products. The cycling parameters were as follows: initial denaturation at 95°C for 5 min; twenty-five cycles of denaturation at 95°C (30 s); annealing at 55°C (30 s); elongation at 72°C (30 s); final extension at 72°C for 5 min. For pyrosequencing, three separate PCR products of each sample were pooled. The PCR products were separated by 1 % agarose gel electrophoresis and purified using the QIAquick Gel Extraction Kit (Qiagen). Amplicons were quantified using the Quant-iT PicoGreen dsDNA Assay Kit (Invitrogen). Equal concentrations of amplicons were pooled from each sample. The amplicons were sequenced using a 454 GS FLX Titanium system (454 Life Sciences/Roche Applied Science) according to the manufacturer's instructions.
Pyrosequencing data analysis
The sequences were processed using the program MOTHUR (version 1.29.0; University of Michigan; http://www.mothur.org/wiki/). The 16S rRNA reads were decoded based on the 5 bp sample-specific barcodes and were processed to remove poor-quality sequences. To reduce sequencing errors, the shhh.flows command was applied, which is the MOTHUR implementation of the AmpliconNoise algorithm( Reference Quince, Lanzen and Davenport 17 ). Quality filters were applied to trim and remove sequences having the following features: length less than 200 bp; average quality score less than 35; homopolymers longer than eight nucleotides; more than two different bases for the primer. To obtain a non-redundant set of sequences, unique sequences were determined and used to align against the SILVA reference alignment database( Reference Pruesse, Quast and Knittel 18 ); chimeras were removed using chimera.uchime (http://drive5.com/uchime); sequences identified to be of eukaryotic origin were removed; the candidate sequences were screened and pre-clustered to eliminate outliers; and a distance matrix was generated from the resulting sequences. The sequences were clustered into operational taxonomic units using the furthest-neighbour algorithm. Representative sequences from operational taxonomic units at a distance of 0·03 were obtained and classified using the SILVA taxonomy and Bayesian classifier( Reference Wang, Garrity and Tiedje 19 ). Rarefaction curves were constructed and Good's coverage values were calculated to quantify the coverage and sampling effort. Community diversity was estimated using the abundance-based coverage estimator (ACE), Chao1 and Shannon indices. The unweighted Unifrac distance method( Reference Lozupone and Knight 20 ) was used to carry out a principal coordinate analysis, and a distance-based analysis of molecular variance (AMOVA) was conducted to assess significant differences between the samples.
Microscopic study
Histological sections were stained with haematoxylin and eosin, embedded in paraffin wax and sectioned as described by Snipes( Reference Snipes 21 ). Additional caecal tissue samples were prepared for electron microscopy using a methodology reported in an earlier study( Reference Snipes 21 ). Washed caecal tissue samples were immediately fixed in 2·5 % glutaraldehyde for 24 h, postfixed for 1 h in 1 % osmium and dehydrated in a graded series of ethanol solutions. For scanning electron microscopy, the tissue samples were subjected to critical-point drying using liquid CO2 as the medium, mounted and coated with gold. The samples were then examined using a scanning electron microscope (Hitachi Model S-3000N, Hitachi Technologies). For transmission electron microscopy, dehydrated samples were placed in a mixture of Spurr resin and acetone (1:1) for 30 min and then in 100 % resin for 10 h. Samples containing resin were placed in moulds and were polymerised at 40 or 60°C for 48 h. Semi-thin (0·25–0·5 μm) sections were cut with glass knives and stained with 1 % toluidine blue O in 1 % sodium borate. Ultra-thin (70–90 nm) sections were cut with a diamond knife, stained with methanolic uranyl acetate and lead citrate, and examined using a transmission electron microscope (Hitachi H-7650, Hitachi Technologies).
Real-time quantitative PCR
Total RNA was extracted from caecal mucosal samples with an acid using the TRIzol reagent (Takara Bio) as described by Chomczynski & Sacchi( Reference Chomczynski and Sacchi 22 ). The concentration of RNA was then quantified using a NanoDrop ND-1000UV–Vis spectrophotometer (Thermo Fisher Scientific). The absorption ratio (260:280 nm) of all the samples was between 1·8 and 2·0, indicating high RNA purity. Aliquots of RNA samples were subjected to electrophoresis on a 1·4 % agarose–formaldehyde gel to verify integrity. The concentration of RNA was adjusted to 1 μg/μl based on optical density, and the samples were stored at − 80°C. Total RNA (1 μg) was reverse-transcribed carefully using the PrimeScript® RT Reagent Kit with gDNA Eraser (Takara Bio) according to the manufacturer's instructions.
The primers for IL-1β, IL-6, TNF-α and interferon-γ (IFN-γ) used have been described by Liu et al. ( Reference Liu, Xu and Liu 1 ) and the primer for glyceraldehyde 3-phosphate dehydrogenase (GAPDH) used has been described by Vorachek et al. ( Reference Vorachek, Bobe and Hall 23 ). Primer sets were designed to recognise and amplify conserved nucleotide sequences encoding goat IL-2 and IL-12. Complementary DNA sequences and/or homologue(s) were identified using the Basic Local Alignment Search Tool computer program (National Center for Biotechnology Information). Primers were designed using the Primer3 computer program (Whitehead Institute). Primers for IL-2 were as follows: forward – 5′-AGTCATTGCTGCTGGATTTACA-3′ and reverse – 5′-CCATTGAATCCTTGATCTCTCTG-3′. Primers for IL-12 were as follows: forward – 5′-GGATCAGAAAGAACCCAAAGC-3′ and reverse – 5′-ATACTCCCTGTGGTCCATGC-3′. All the primers were synthesised by Invitrogen Life Technologies. Real-time quantitative PCR of target genes and GAPDH were carried out using the ABI 7300 Real-Time PCR System (Applied Biosystems) through fluorescence detection using the SYBR Green dye. Amplification conditions were as follows: 95°C for 30 s, followed by forty cycles comprising 5 s at 95°C and 31 s at 57·5°C (for GAPDH) or at 60°C (for other genes). Each sample contained 1–10 ng of complementary DNA in 2 × SYBR Green PCR Master Mix (Takara Bio) and 200 nmol/l of each primer in a final volume of 20 μl. All measurements were made in triplicate. A reverse-transcription-negative blank of each sample and a no-template blank were used as negative controls. The relative amount of each studied mRNA was normalised to that of GAPDH mRNA used as a housekeeping gene, and the data were analysed according to the 2− ΔΔC T method.
Data analysis
Statistical calculations were carried out by conducting tests using the SPSS software package (SPSS version 16, SPSS, Inc.). The normality of the distribution of variables was tested using the Shapiro–Wilk test. The independent-samples t test procedure was used to analyse variables found to have a normal distribution. The Kruskal–Wallis test was used to analyse variables found to have a non-normal distribution. Significance was set at P< 0·05.
Double dendrograms were constructed using the comparative functions and multivariate hierarchical clustering methods of NCSS 2007 (NCSS), on the basis of the abundance of bacterial groups at different taxonomic levels. Clustering was done using Ward's minimum variance method with no scaling. Correlations between caecal pH, caecal LPS concentration, and mucosa-associated microbiota abundance and inflammatory cytokine expression were assessed by Pearson's correlation test using GraphPad Prism version 5.00 (GraphPad Software). Significance was set at P< 0·05.
Results
Ruminal pH and concentrations of volatile fatty acids, lactate and lipopolysaccharide
The results reported herein need to be interpreted in relation to the overall effects of HG feeding on the rumen fermentation of experimental goats reported previously( Reference Liu, Xu and Liu 1 ) (Table S2, available online). Briefly, the ruminal pH of the HG diet-fed goats was lower (P< 0·001) than that of the hay diet-fed goats. HG feeding increased (P< 0·001 to P< 0·019) the concentrations of propionate, butyrate, valerate, isovalerate, total VFA and lactate.
Caecal digesta pH and concentrations of volatile fatty acids, lactate, lipopolysaccharide and starch
Compared with the hay diet, the HG diet decreased the caecal digesta pH (P< 0·001) and concentration of isobutyrate (P= 0·01) and increased the concentrations of acetate (P< 0·001), propionate (P< 0·001), butyrate (P= 0·002) and total VFA (P< 0·001) (Table 1). No significant difference was observed in the concentrations of valerate (P= 0·663), isovalerate (P= 0·328) and lactate (P= 0·922) between the control and HG groups (Table 1). The proportion of starch in the caecal digesta of the HG group was 0·66 %. The concentration of free LPS in the caecal digesta was significantly increased (P= 0·004) during HG feeding.
Table 1 Effects of high-grain feeding on caecal physiological parameters in goats at the time of slaughter (Mean values with their standard errors; n 5)

VFA, volatile fatty acids; ND, not detectable; LPS, lipopolysaccharide; EU, endotoxin unit.
General DNA sequencing observations
In total, 65 368 and 58 010 reads were obtained for the 16S rRNA gene in the caecal luminal content and mucosa of all goats and 56 346 and 53 114 were valid correspondingly, accounting for 86·04 and 91·71 % of their raw reads (Table S3, available online). The sequences were further analysed using the MOTHUR software, and a total of twenty-three phyla were detected. Of these, sixteen phyla were detected in the caecal luminal contents of the hay diet-fed group and sixteen phyla in those of the HG diet-fed group and nineteen phyla were detected in the mucosal epithelium of the hay diet-fed group and twenty-two phyla in that of the HG diet-fed group. At the genus level, these sequences were assigned to 362 different genera. Among these, 112 and ninety-four genera were detected in the caecal luminal content and mucosa across all the goats, respectively, and 159 and 314 genera were detected in the mucosal epithelium of the hay diet-fed and HG diet-fed groups, respectively.
Diversity of caecal microbiota
The rarefaction curves of caecal bacterial communities are shown in Fig. S1 (available online). At dissimilarity levels of 0·03 (‘species’), 0·05 (‘genus’) and 0·10 (‘family’), the rarefaction analysis revealed that HG feeding decreased bacterial diversity in the caecal lumen compared with the control (Fig. S1(A), (C) and (E), available online). However, there was no apparent difference in bacterial diversity in the caecal mucosa between the two groups (Fig. S1(B), (D) and (F), available online). At the 0·03 dissimilarity level, the hay diet-fed group exhibited significantly high microbial richness in the caecal lumen when compared with the HG diet-fed group (P< 0·001) (Table 2). Analysis based on the Shannon index indicated that the Shannon indices of the HG diet-fed group were lower (P< 0·001) than those of the hay diet-fed group. However, no significant changes were observed (P>0·10) in the operational taxonomic units and ACE, Chao1 and Shannon indices in the caecal mucosa between the hay diet-fed and HG diet-fed groups.
Table 2 Effects of high-grain feeding on the diversity of caecal bacterial community at the 3 % dissimilarity level (Mean values with their standard errors; n 5)
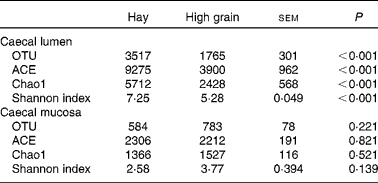
OTU, operational taxonomic units; ACE, abundance-based coverage estimator.
To compare community compositions across the samples, we used the unweighted UniFrac metric in MOTHUR to assess β-diversity. The results indicated that unweighted UniFrac were able to distinctly separate all the control samples from the HG samples of the caecal lumen (Fig. 1(a)) and caecal mucosa (Fig. 1(b)). Principal coordinate analysis with unweighted UniFrac revealed that principal component analysis (PCA) axis 1 accounted for 65·0 % (lumen) or 18·6 % (mucosa) of the variation and PCA axis 2 for 11·1 or 13·6 % of the variation, respectively. In addition, we found that bacterial communities in the caecal lumen (AMOVA; F s= 12·42, P= 0·006) and caecal mucosa (AMOVA; F s= 10·73, P= 0·007) were significantly affected by diet.

Fig. 1 Results of the principal coordinate analysis (PCoA) of bacterial communities in the caecal luminal or mucosal samples. (a) Unweighted PCoA by caecal luminal microbiota; (b) unweighted PCoA by caecal mucosa-associated microbiota; (c) distribution of the phyla averaged across the dietary treatments in the caecal luminal samples; (d) distribution of the phyla averaged across the dietary treatments in the caecal mucosal samples. Hay-L, luminal sample from goats fed the hay diet; HG-L, luminal sample from goats fed the high-grain diet; hay-M, mucosal sample from goats fed the hay diet; HG-M, mucosal sample from goats fed the high-grain diet; PC, principal coordinates. (a) ![]() , Hay;
, Hay; ![]() , high-grain. (b)
, high-grain. (b) ![]() , hay;
, hay; ![]() , high-grain. (c)
, high-grain. (c) ![]() , Acidobacteria;
, Acidobacteria; ![]() , Deferribacteres;
, Deferribacteres; ![]() , Synergistetes;
, Synergistetes; ![]() , Elusimicrobia;
, Elusimicrobia; ![]() , Fibrobacteres;
, Fibrobacteres; ![]() , Cyanobacteria;
, Cyanobacteria; ![]() , Chloroflexi;
, Chloroflexi; ![]() , Verrucomicrobia;
, Verrucomicrobia; ![]() , Lentisphaerae;
, Lentisphaerae; ![]() , Planctomycetes;
, Planctomycetes; ![]() , Proteobacteria;
, Proteobacteria; ![]() , Spirochaetes;
, Spirochaetes; ![]() , Tenericutes;
, Tenericutes; ![]() , Actinobacteria;
, Actinobacteria; ![]() , unclassified Bacteria;
, unclassified Bacteria; ![]() , Bacteroidetes;
, Bacteroidetes; ![]() , Firmicutes. (d)
, Firmicutes. (d) ![]() , Elusimicrobia;
, Elusimicrobia; ![]() , Deinococcus-Thermus;
, Deinococcus-Thermus; ![]() , Armatimonadetes;
, Armatimonadetes; ![]() , Gemmatimonadetes;
, Gemmatimonadetes; ![]() , Chlorobi;
, Chlorobi; ![]() , Verrucomicrobia;
, Verrucomicrobia; ![]() , Fusobacteria;
, Fusobacteria; ![]() , Nitrospirae;
, Nitrospirae; ![]() , Cyanobacteria;
, Cyanobacteria; ![]() , Tenericutes;
, Tenericutes; ![]() , Acidobacteria;
, Acidobacteria; ![]() , Chloroflexi;
, Chloroflexi; ![]() , Fibrobacteres;
, Fibrobacteres; ![]() , Planctomycetes;
, Planctomycetes; ![]() , Lentisphaerae;
, Lentisphaerae; ![]() , Unclassified Bacteria;
, Unclassified Bacteria; ![]() , Actinobacteria;
, Actinobacteria; ![]() , Bacteroidetes;
, Bacteroidetes; ![]() , Firmicutes;
, Firmicutes; ![]() , Proteobacteria;
, Proteobacteria; ![]() , Deferribacteres;
, Deferribacteres; ![]() , Spirochaetes.
, Spirochaetes.
Composition of caecal microbiota
Compared with microbiota in the caecal mucosa, the phylum Firmicutes had an absolute advantage with regard to abundance in the caecal luminal content, accounting for more than 75 % of total sequences (Fig. S2, available online). However, the phylum Spirochaetes exhibited a considerable abundance in most of the mucosal samples (Fig. S2, available online). At the phylum level, the phyla Bacteroidetes and Firmicutes together represented more than 90 % of the microbial community in the caecal luminal content in the two dietary groups. Among all the sixteen phyla detected in the caecal luminal samples (Fig. 1(c)), the abundance of only three bacterial phyla including Actinobacteria, Spirochaetes and Chloroflexi was significantly affected by diet (P< 0·01). Among these, HG feeding decreased (P< 0·01) the abundance of the phyla Actinobacteria and Chloroflexi and increased (P< 0·01) that of the phylum Spirochaetes. In the caecal mucosa samples (Fig. 1(d)), when compared with the control, HG feeding increased (P= 0·008 to P= 0·032) the abundance of the phyla Planctomycetes, Fibrobacteres, Nitrospirae, Verrucomicrobia, Armatimonadetes, Elusimicrobia, Firmicutes, Bacteroidetes, Chloroflexi, Acidobacteria and Deinococcus-Thermus and decreased that of the phylum Deferribacteres (P= 0·008).
For clarity and visualisation purposes, the top fifty bacterial genera are presented in a heat map. The heat map analysis of the caecal luminal samples revealed that Clostridium, Turicibacter, unclassified Rikenellaceae, unclassified Peptostreptococcaceae, unclassified Clostridiales, unclassified Lachnospiraceae and unclassified Ruminococcaceae occurred together in one cluster, while the other forty-three genera occurred in another main cluster (Fig. S3, available online). In Table S4 (available online), the predominant genera (the average relative abundance >0·5 %) in the caecal content significantly affected (P< 0·05) by HG feeding are listed. In general, when compared with the control, HG feeding increased (P= 0·002 to P= 0·009) the proportion of Treponema, Turicibacter, Prevotella, Clostridium and unclassified Firmicutes and decreased (P< 0·001 to P< 0·015) the proportion of Bacteroides, Phocaeicola, Phascolarctobacterium and some unclassified Bacteria including unclassified Ruminococcaceae, unclassified Peptostreptococcaceae, unclassified Coriobacteriaceae, unclassified Prevotellaceae, unclassified Clostridiales and unclassified Lachnospiraceae.
In the mucosal samples, among the top fifty genera detected, the genus Treponema occurred in one cluster and the other forty-nine genera occurred in another main cluster (Fig. S4, available online). When compared with the control, HG feeding increased (P< 0·001 to P< 0·031) the proportion of Turicibacter, Clostridium, Mogibacterium, Bacteroides, Ruminococcus, Nitrospiraceae_Nitrospira and some unclassified taxa including unclassified Bacteroidales, unclassified Gammaproteobacteria, unclassified Rikenellaceae, unclassified Fibrobacteres and unclassified Rhodocyclaceae and decreased (P= 0·009) the proportion of Mucispirillum. (Table S5, available online).
Histology, morphology and ultrastructure of caecal epithelium
Representative light micrographs of the cross sections of caecal epithelium of the hay diet-fed and HG diet-fed goats are shown in Fig. S5(A) and (B) (available online). Scanning electron micrographs of the caecal epithelium of the hay diet-fed and HG diet-fed goats are shown in Fig. S5(C) and (D) (available online). In the hay diet-fed goats, the orifices of crypts were circular in outline and the intercryptal surface was partially covered by an irregular layer of mucus (Fig. S5(A) and (C), available online). In contrast, the HG diet-fed goats exhibited lower crypt density and sloughing of the epithelial surface (Fig. S5(B) and (D), available online).
Transmission electron micrographs of the cross sections of caecal epithelia were generated (Fig. S6, available online). The hay diet-fed goats had clear and organised microvillus clusters (Fig. S6(A), available online). However, the microvilli of the HG diet-fed goats were sparse and messy (Fig. S6(B), available online). In the hay diet-fed goats, the integrity of cell organelles in the microvilli was normal (Fig. S6(A), available online), and the tight junction band was clearly visible (Fig. S6(C), available online). In the HG diet-fed goats, numerous vacuoles appeared in the cell layers. Mitochondrial swelling (Fig. S6(B), available online) and intercellular tight junction erosion were also apparent (Fig. S6(D), available online).
Relative mRNA expression of cytokines in the caecal tissue
The mRNA expression levels of cytokines, as measured by quantitative RT-PCR, are shown in Fig. 2. The data revealed that there was no significant difference in the expression of IL-2 (P= 0·260) and TNF-α (P= 0·065) genes in the caecal tissue between the hay diet-fed and HG diet-fed goats. Compared with the hay diet, the HG diet increased the relative mRNA expression of IL-1β (P< 0·001), IL-6 (P= 0·006), IL-12 (P< 0·001) and IFN-γ (P= 0·025).

Fig. 2 Effects of high-grain (■) feeding on the relative mRNA expression of cytokines in the caecum of goats. Values are means (n 5), with their standard errors represented by vertical bars. Mean value was significantly different from that of the hay (![]() ) diet-fed group: *P< 0·05; **P< 0·01; ***P< 0·001.
) diet-fed group: *P< 0·05; **P< 0·01; ***P< 0·001.
Correlation analysis
A correlation analysis was carried out to determine whether there were any relationships between mucosal inflammatory cytokine expression and luminal pH, LPS concentration and mucosa-associated microbiota abundance. In Fig. 3, the relationship between the abundance of the predominant bacterial genera (relative abundance ≥ 1 % in at least one sample) and the expression of inflammatory cytokines is shown. The relative mRNA expression of IL-6 (r − 0·908; P< 0·001) and IL-12 (r − 0·855; P= 0·002) in the mucosa was negatively correlated with the luminal pH. The concentration of LPS was positively correlated with the relative mRNA expression of IL-6 (r 0·641; P= 0·046) and IL-12 (r 0·689; P= 0·028) in the mucosa. In addition, the abundance of three genera, including Anoxybacillus, Silanimonas and Brevibacillus, was positively correlated (P< 0·05) with the relative mRNA expression of IL-2; the abundance of two taxa (Turicibacter and unclassified Bacteroidales) was positively associated (P< 0·05) with the relative mRNA expression of IL-6; the abundance of eleven taxa was correlated (P< 0·05) with the relative mRNA expression of IL-12 (one negative (Mucispirillum) and ten positive (Turicibacter, unclassified Bacteroidales, unclassified Bacteria, Clostridium, Nitrospiraceae_Nitrospira, unclassified Gammaproteobacteria, unclassified Ruminococcaceae, unclassified Clostridiales, unclassified Fibrobacteres and Alistipes)); the abundance of four taxa (Clostridium, unclassified Ruminococcaceae, unclassified Clostridiales and unclassified Fibrobacteres) was positively associated (P< 0·05) with the relative mRNA expression of TNF-α and that of eleven taxa (Clostridium, unclassified Ruminococcaceae, unclassified Bacteria, Nitrospiraceae_Nitrospira, unclassified Gammaproteobacteria, Anoxybacillus, Silanimonas, Brevibacillus, unclassified Rhizobiales, unclassified Rhodocyclaceae and Marinomonas) was positively associated (P< 0·05) with the relative mRNA expression of IFN-γ.

Fig. 3 Results of the correlation analysis between the caecal pH, caecal lipopolysaccharide (LPS) concentration and mucosa-associated microbiota composition (at the genus level) and the expression levels of mucosal inflammatory cytokines. Only results obtained for the predominant bacterial genera (relative abundance ≥ 1 % in at least one sample) for which the abundance was significantly associated with inflammatory cytokine expression are shown. Cells are coloured based on Pearson's correlation coefficient between the caecum pH, caecal LPS concentration, and mucosa-associated microbiota composition and the expression levels of inflammatory cytokines in the caecal mucosa. Red colour represents a significant positive correlation (P< 0·05), blue colour represents a significant negative correlation (P< 0·05), and green colour represent a non-significant correlation (P>0·05).
Discussion
Alterations in caecal fermentation during high-grain feeding
Over the past 50 years, meat and milk production per goat or dairy cattle has increased dramatically. One reason for this increase is the feeding rations that have greater proportions of grains and lower proportions of forages. Higher proportions of grains lead to higher production of VFA and increased energy supply to the host( Reference Li, Khafipour and Krause 7 , Reference Clark and Davis 24 ). In an experiment conducted concurrently with the present study, as described in the paper of Liu et al. ( Reference Liu, Xu and Liu 1 ), the concentrations of total VFA in the HG group were 17·2 % higher than those in the control group, and this indicates that a HG diet increases rumen microbial fermentation and results in higher VFA production in the rumen. These results were also consistent with the findings mentioned earlier. However, a high-concentrate diet has been found to reduce the production of saliva and bicarbonate, which buffer the rumen( Reference Owens, Secrist and Hill 25 ). A reduced buffering capacity, combined with increased VFA production, has been reported to decrease ruminal pH and increase acidosis incidence( Reference Owens, Secrist and Hill 25 ). Acidosis often results in an increased amount of fermentable carbohydrates passing to the hindgut, thus increasing the fermentation in the hindgut( Reference Gressley, Hall and Armentano 4 , Reference Li, Khafipour and Krause 7 ). The increase in starch concentration in the caecal digesta due to the HG diet observed in the present study confirms the findings of many earlier studies that showed that increasing dietary starch content will increase the proportion of the undigested starch that bypasses fermentation in the rumen and digestion in the small intestine( Reference Huntington 26 , Reference Owens, Zinn and Kim 27 ). As in the rumen, increased starch concentrations in the hindgut will increase the production of VFA and decrease the pH of the digesta( Reference Li, Khafipour and Krause 7 , Reference Owens, Zinn and Kim 27 ). In the present study, the HG diet decreased the caecal pH and increased the caecal total VFA concentrations. These results were consistent with the reports mentioned earlier and suggest that HG diets lead to an increase in fermentation in the large intestine. However, the caecal pH of the HG group in the present study was lower than that of the cattle fed a HG diet in the study of Li et al. ( Reference Li, Khafipour and Krause 7 ). This could be expected, because the diets used in the present study contained a greater proportion of grains than the grain-based subacute ruminal acidosis challenge diets used in the study of Li et al. ( Reference Li, Khafipour and Krause 7 ).
Global alterations in the microbial community structure during high-grain feeding
Compared with the advanced understanding that exists about caprine rumen microbiota and of its adaptation to different dietary regimens, alterations in the goat caecal microbial community in response to dietary changes are less well characterised. Therefore, the present study is the first (to our knowledge) to compare the adaptive responses of the caecal microbiota to HG feeding using high-throughput sequencing methods. In line with previous findings reported for the colon microbial communities of steers( Reference Romero-Perez, Ominski and McAllister 28 ) and cattle( Reference Godoy-Vitorino, Goldfarb and Karaoz 29 ), members of the phylum Firmicutes were the most abundant and those of the phylum Bacteroidetes were the second most abundant luminal bacterial populations investigated in the present study, highlighting their importance in polysaccharide degradation. In contrast, members of the phylum Spirochaetes, which are part of the normal intestinal flora and have been isolated from the gut mucosa of humans and cattle( Reference Lu and Walker 30 , Reference Malmuthuge, Griebel and Guan le 31 ), are highly abundant in the caecal mucosa. It is likely that these differences between the composition of the luminal microbiota and that of the mucosa-associated microbiota may be due to the difference in ecology niches of microbial and nutrient sources available for microbial growth( Reference Lu and Walker 30 ). However, the abundance of members of the phylum Spirochaetes observed in the present study was higher than that in the caecum mucosa of calves fed whole milk( Reference Malmuthuge, Griebel and Guan le 31 ). It is possible that this difference was due to differences in the diet composition and host phylogeny, because it is known that the composition of the gut microbial community is related to the host phylogeny and diet type( Reference Zhang, Zhang and Wang 32 ). In addition to these, there were also a number of phyla that are not generally considered to be gut microbes, including Deinococcus-Thermus and Cyanobacteria. Of these, Cyanobacteria had been detected previously in human and other animal guts( Reference Di Rienzi, Sharon and Wrighton 33 ). Ley et al. ( Reference Ley, Backhed and Turnbaugh 34 ) reported that there is a coherent gut-associated clade rooted deep in the Cyanobacteria group, and this group may represent descendants of non-photosynthetic ancestral cyanobacteria that have adapted to life in animal gastrointestinal tracts. Deinococcus-related organisms are largely associated with extreme environments, but have recently been identified in the human stomach( Reference Delgado, Cabrera-Rubio and Mira 35 ) and in appendices( Reference Guinane, Tadrous and Fouhy 36 ). The presence of these phyla indicates that environmentally resistant organisms reside within the caecum of goats. The increase in the abundance of Deinococcus-Thermus in the caecal mucosa of the HG group observed in the present study indicates that HG feeding may result in an unhealthy mucosal environment and favour the growth of more environmentally resistant organisms in the caecal mucosa of goats.
The principal coordinate analysis and AMOVA revealed clear differences in the composition of the microbial community in the caecal lumen between the control and HG groups. These results indicate that the structure of the caecal lumen microbiota in the HG diet-fed group was different from the structure of those in the control group. This difference in the microbial population structure may be due to the increased amounts of fermentable substrates present in the diet favouring the growth of amylolytic and other starch-digesting bacterial species. Indeed, the present study reveals that a HG diet supports a higher proportion of members of several genera in the caecal luminal contents, including Turicibacter, Clostridium, Treponema and Prevotella. Of these, Turicibacter is a relatively unknown genus. Recent reports of 16S rRNA gene and ribosomal intergenic spacer analysis data indicate the presence of Turicibacter bacteria in the rumen and faeces of cattle( Reference Mao, Zhang and Wang 37 , Reference Shanks, Kelty and Archibeque 38 ). Turicibacter has also been reported to be present in pig, rat and insect hindguts, dairy wastewaters and raw milk( Reference Ahn, Hong and Bok 39 – Reference Gagnon, Talbot and Ward 41 ). However, as only one species (Turicibacter sanguinis) has been cultured( Reference Bosshard, Zbinden and Altwegg 42 ), the physiological diversity of the members of this genus is unknown. As the isolated strain is a putative pathogen, it is possible that Turicibacter bacteria, present in goats, may cause subclinical infection or have some other deleterious effect on the gastrointestinal tract. Clostridium is well known as a gut coloniser, which is commonly found in the intestine of goats( Reference Garcia, Adams and Beingesser 43 ). Clostridium genus comprises some opportunistic pathogens such as Clostridium difficile whose increased intestinal proliferation is critically regarded as a causal agent of enteric diseases in goats( Reference Garcia, Adams and Beingesser 43 ). Azcarate-Peril et al. ( Reference Azcarate-Peril, Foster and Cadenas 44 ) also reported that there is a higher abundance of Clostridium spp. in the ileal mucosa of pigs with necrotising enterocolitis than in that of healthy pigs. Thus, these findings imply that the increase in the abundance of Clostridium and Treponema during HG feeding could have some deleterious effects on the caecal health. In the present study, the cellulolytic bacteria Lachnospiraceae, Ruminococcaceae and Clostridiales were found in higher abundance in the lumen of the hay diet-fed group than in that of the HG diet-fed group. This is consistent with the findings recorded in the rumen of dairy cattle fed a high-forage diet( Reference McLellan, Newton and Vandewalle 45 ). In addition, HG feeding also altered the composition of the mucosa-associated microbial ecosystem, and this alteration may be caused by the changes in the growth substrate of caecal microbiota. During forage feeding, the mucosa-associated microbiota mainly used mucus as a growth substrate. However, diet-derived nutrients such as starch are likely to be used by mucosa-associated bacteria during HG feeding, thus resulting in an increase in the proportion of polysaccharide-degrading bacteria such as Clostridium and Treponema ( Reference Tajima, Aminov and Nagamine 46 ). In contrast, the proportion of Mucispirillum, which has an affinity for mucosal surfaces( Reference Robertson, O'Rourke and Neilan 47 ), was decreased in the HG diet-fed group. This might be partly due to an increased proportion of the polysaccharide-degrading bacteria in the caecal mucosa. Another possible reason for the decrease in the proportion of Mucispirillum observed in the present study may be related to the lower pH of digesta resulting from HG feeding, because Gram-negative bacteria are known to be sensitive to a low pH( Reference Tajima, Aminov and Nagamine 46 ).
The major impact of HG feeding on the activity of the caecal microflora in goats can observed in the production of fermentation products, mainly three monocarboxylates commonly referred to as VFA, acetate, propionate and butyrate. As has been mentioned earlier, in the present study, HG feeding decreased the caecal pH and increased the caecal total VFA concentrations. Interestingly, in the present study, about 3-fold increase in the concentration of butyrate was observed in the HG group compared with the control group. This could be explained by the higher abundance of Clostridium observed in the lumen of the HG diet-fed group, as Clostridium spp. have been considered to be the predominant butyrate-producing bacteria in the human gut lumen( Reference Van den Abbeele, Belzer and Goossens 48 ). In addition, the correlation analysis also revealed that there was a significant positive correlation between the abundance of Clostridium and the concentration of butyrate (R 0·954; P< 0·001, data not shown). Thus, the higher concentration of butyrate in the caecal digesta may be partly due to the fermentation activity of the increased proportion of Clostridium spp. in the HG group.
LPS is a structural component of the outer membrane of Gram-negative bacteria. Although LPS is bound to the outer membrane, it is shed during the growth and stationary phases of bacterial growth and is released after cell disintegration and lysis( Reference Nagaraja, Bartley and Fina 49 ). Several studies have shown that feeding cattle or goats with a HG diet will increase the concentration of LPS in the rumen( Reference Liu, Xu and Liu 1 , Reference Nagaraja, Bartley and Fina 49 , Reference Gozho, Plaizier and Krause 50 ). Nevertheless, Bertok( Reference Bertok 51 ) found that bile acids cause the degradation of LPS in the small intestine, and Ribeiro et al. ( Reference Ribeiro, Xu and Klein 52 ) reported that the acidic conditions in the abomasum also deactivate LPS. Hence, it can be speculated that the increase in LPS concentration in the caecum of the HG group is due to the increased growth and (or) lysis of LPS-producing bacteria in the hindgut but not in the rumen. It has been well established that a decrease in the ruminal pH of dairy cows fed a HG diet is associated with a significant increase in the concentration of LPS in the rumen and that this is usually accompanied by a decrease in the abundance of Gram-negative bacteria in the rumen( Reference Plaizier, Khafipour and Li 12 , Reference Nagaraja, Bartley and Fina 49 ). In the present study, significant negative correlations were observed between the concentration of free LPS in the caecal digesta and the proportion of Gram-negative bacteria, including the taxa Bacteroides (R − 0·90; P< 0·001), unclassified Prevotellaceae (R − 0·70; P= 0·026), Phascolarctobacterium (R − 0·68; P= 0·030) and Phocaeicola (R − 0·67; P= 0·033, data not shown). Moreover, the abundance of the taxa mentioned earlier was much lower in the HG group than in the control group (Table S4, available online). These findings demonstrate that HG feeding results in a low caecal pH and that the latter could lead to the death and lysis of some Gram-negative bacteria, thereby increasing the concentration of free LPS in the caecum.
Mucosal injury and its correlation with luminal pH, lipopolysaccharide concentration and mucus-associated microbiota abundance
In the present study, after 7 weeks of feeding, the HG diet-fed goats exhibited profound alterations in caecal epithelial structure, depicted by sloughing of epithelial surface, lower crypt density, intercellular tight junction erosion and cell necrosis. In addition, the expression of inflammation cytokines, including IL-1β, IL-6, IL-12 and IFN-γ, was up-regulated in the HG group compared with that in the control group. These findings, for the first time, demonstrate that a HG diet causes profound caecal epithelial cellular damage, which is associated with a local inflammatory response in the caecal epithelium. There are many possible explanations for the mucosal injury observed in the HG group. Excessive amounts of carbohydrates in the caecum of the HG diet-fed goats may increase the osmotic pressure of digesta. High osmotic pressure is undoubtedly a threat to the epithelia barrier and may increase the permeability of caecal epithelia( Reference Gressley, Hall and Armentano 4 ). LPS and acidic pH are known to increase the permeability of the mucosa independently( Reference Chin, Flynn and Fedwick 11 ). Therefore, these findings together with the investigation of inflammation cytokine expression in the mucosa indicate that the increase in acidity, coupled with LPS, may work to damage the caecal mucosal epithelium, thereby initiating a local inflammatory response.
In spite of the present study revealing the detrimental effects of a HG diet on the integrity of the intestinal epithelium, the principle mechanisms responsible for triggering a local immune response during HG feeding are unknown. The correlation analysis carried out in the present study revealed that there is a relationship between LPS or pH and some inflammation cytokines such as IL-6 and IL-12. These results indicate that the higher LPS concentrations and lower pH may partly contribute to the increased local inflammation in the HG group. Indeed, the influence of a lower pH on inflammation is well documented in the colon and is probably the mechanism that triggers the inflammatory response in the caecal epithelium( Reference Mortensen and Clausen 53 ). It is well known that VFA have high lipid solubility. When environmental pH declines to the threshold level, VFA can enter the non-glandular mucosal cells, acidifying the cells and damaging chloride-dependent Na transport, which results in cell necrosis( Reference Andrews, Buchanan and Elliot 54 ). Additionally, the endotoxic effects of LPS on the caecal epithelium may be partly responsible for the initial local inflammatory reaction, leading to an immune-mediated cascade of events to repair the caecal epithelium( Reference Thibault, Blachier and Darcy-Vrillon 55 ).
It should be stressed that not only lower pH and LPS but also some pathogenic and opportunistic pathogenic bacteria can induce inflammation. Previous studies have revealed that intestinal mucosal tissue damage in colitis may mostly result from the production of inflammatory cytokines in response to commensal bacteria( Reference Saleh and Trinchieri 56 , Reference Elinav, Strowig and Kau 57 ). Correlation analysis of the mucosa-associated microbiota composition and inflammatory parameters carried out in the present study revealed a correlation between the increase in the expression of some pro-inflammatory cytokines and the changes in the abundance of some mucosal microbial populations in goats. In particular, increases in the expression of IL-6 and IL-12, which was observed in the HG diet-fed goats, were associated with the enrichment of Turicibacter and Clostridium populations in the HG group. As has been mentioned earlier, some of the Turicibacter and Clostridium phylotypes are known to have deleterious effects leading to mucosal injury( Reference Kellermayer, Dowd and Harris 58 , Reference Lupp, Robertson and Wickham 59 ). Hence, these results reveal that the inflammation that occurs in the hindgut mucosal epithelium may partly arise from these deleterious effects.
The correlation analysis carried out in the present study also revealed that 122 genera (mucosa-associated microbiota) were not linked to the expression of inflammation cytokines evaluated (data not shown), and this indicates that there are fractions of the resident mucosa-associated microbiota that are less likely to induce inflammation. In contrast, in the present study, it was found that the increase in the expression of IL-12, which was observed in the HG diet-fed goats, was linked to a decrease in the abundance of Mucispirillum spp. (Fig. 3). These results indicate that Mucispirillum may have the ability to counteract inflammation. This seemingly inflammation-suppressing effect can be a result of Mucispirillum occupying the adhesion sites on the mucosa during forage feeding and thereby preventing the invasion of some inflammation-aggravating bacteria, which will decrease the inflammatory tone of the system. Nevertheless, it should be emphasised that although a significant alteration in Mucispirillum composition and its association with the decrease in the expression of some inflammation cytokines strongly indicate that some mucosa-associated microbial populations might have anti-inflammatory effects, a direct experimental demonstration of the anti-inflammatory effects of Mucispirillum has not been reported. Thus, a direct demonstration experiment to investigate whether Mucispirillum spp. colonisation protects animals from inflammatory disease is urgently needed to confirm these hypotheses.
There has been remarkably little information concerning the effects of caecal epithelial damage on caecum function and animal performance. However, recent studies on the influence of rumen acidosis on rumen function have highlighted the possibility that caecal epithelial injury has a significant impact on caecum function. Previous studies have revealed that rumen acidosis may increase the permeability of the rumen epithelium( Reference Liu, Xu and Liu 1 , Reference Khafipour, Krause and Plaizier 60 ) and that this increased permeability may increase the uptake of toxic compounds such as histamine generated in the rumen in the body and the uptake of VFA( Reference Wang, Zhang and Zhu 61 ). In addition, chronic inflammation caused by ruminal epithelial damage may stimulate the release of cytokines and trigger an immune response( Reference Khafipour, Krause and Plaizier 60 ), and the latter may decrease feed intake and diet digestibility( Reference Plaizier, Krause and Gozho 62 ). Furthermore, some recent studies have revealed that hindgut acidosis decreases diet digestibility and milk fat percentage in cattle, and this decreased diet digestibility probably results from increased bulk in the digestive tract or from increased digesta passage rate, reducing the exposure of digesta to intestinal enzymes and epithelial absorptive surfaces( Reference Gressley, Hall and Armentano 4 , Reference Mainardi, Hengst and Nebzydoski 63 ). The permeability of caecal epithelium and diet digestibility in the caecum were not measured in the present study. However, Emmanuel et al. ( Reference Emmanuel, Madsen and Churchill 10 ) demonstrated that a low pH in the presence of LPS increases the permeability of the colon epithelium of cattle. Therefore, these findings indicate that the damage caused to the caecal epithelium by HG feeding may have a potentially adverse impact on caecum function and animal performance.
Conclusions
Data obtained in the present study revealed that HG feeding caused a decrease in the caecal pH and an increase in the concentrations of total caecal VFA, lactic acid and LPS, indicating increased fermentation and production of free LPS in the large intestine of goats. They also revealed that the response of bacterial populations in the caecal lumen and mucosa of goats was associated with HG feeding. The greatest shifts in the predominant bacterial populations were observed for caecal luminal Turicibacter, Clostridium and unclassified Ruminococcaceae and mucosa-associated Mucispirillum spp., Turicibacter spp., Clostridium spp., Gammaproteobacteria spp. and Rikenellaceae spp. in goats fed the HG diet compared with those fed the hay diet, indicating that the substrate flow into the large intestine alters the abundance of bacterial populations. Microscopic examinations of the caecal mucosal samples revealed that HG feeding caused mucosal epithelial injury and RT-PCR revealed that the mucosal injury might be caused by the up-regulated expression of IL-6 and IL-12 in the caecal mucosa. The correlation analysis revealed that alterations in caecal pH, LPS concentration and mucosa-associated microbiota composition during HG feeding might partly contribute to local inflammation. Overall, these results provide a comprehensive picture of the caecal microbial structure during HG feeding and help to further elucidate the aetiology of hindgut disorders. However, more detailed information concerning mucosa-associated microbiota and its relationship with inflammation is required. Future work should focus on the isolation of anti-inflammatory species for the prevention of mucosal injury and the modulation of intestinal epithelial restitution in the hindgut as a response to HG feeding in goats.
Supplementary material
To view supplementary material for this article, please visit http://dx.doi.org/10.1017/S0007114514000993
Acknowledgements
The present study was supported by the National Natural Science Foundation of China (no. 31372339 and 31172228).
The authors' contributions are as follows: J. L. and T. X. carried out the majority of the animal studies including animal care, VFA analysis, RNA isolation and real-time PCR; S. M. carried out microbial data processing, analysis and interpretation; S. M. and W. Z. were responsible for the conception of the project and for overseeing the experiment; J. L. and S. M. were responsible for writing the manuscript.
None of the authors has any conflicts of interest to declare.







