Obesity is a multifactorial disorder reflecting complex interactions of genes, environment and lifestyle(Reference Newell, Zlot and Silvey1). Industrialisation and modernisation encourage a sedentary lifestyle with concomitantly increased energy intake, resulting in an imbalance of energy intake and expenditure(Reference Bell, Walley and Froguel2) and consequently in gaining surplus weight. About 40–70 % of the variance in the BMI is accounted for by genetic factors as several studies of twins, adoptees and families have shown(Reference Atwood, Heard-Costa and Cupples3–Reference Salsberry and Reagan5). Screenings of candidate regions as well as genome-wide scans have helped to identify SNP that increase the risk of becoming overweight or obese(Reference Peeters, Beckers and Verrijken6).
There is considerable evidence that not all fatty acids are obesogenic(Reference Storlien, Huang and Lin7). Approximately 6·2–7·4 % of our average daily energy intake is accounted for by PUFA (n-3 PUFA 0·7–0·9 %; n-6 PUFA 5·5–6·5 %)(Reference Linseisen, Schulze and Saadatian-Elahi8). PUFA exert their influence on cardiovascular function, insulin action, plasma lipid levels(Reference Harris, Lu and Rambjor9), neuronal development and the immune system inter alia through modulation of eicosanoid (PG and leukotrine) synthesis, activation of orphan nuclear receptor and T-lymphocyte signalling(Reference Harris, Lu and Rambjor9, Reference Stulnig10). They also regulate the transcription and activation of multiple genes(Reference Jump11–Reference Sampath and Ntambi13). Hence, PUFA affect several metabolic pathways, and thus may have an impact on the development of a series of diseases, including obesity(Reference Sampath and Ntambi13, Reference Storlien, Hulbert and Else14) either directly or through interactive effects with the genetic background. This is supported by the results of different experimental studies(Reference Iwaki, Matsuda and Maeda15–Reference Verlengia, Gorjao and Kanunfre18).
So far, only a few studies have investigated gene–PUFA interactions with respect to obesity risk. We have mainly found human or animal cell studies or dietary intervention studies showing PUFA to have an increasing or decreasing effect on the expression of different candidate genes for obesity. Reseland et al. (Reference Reseland, Haugen and Hollung17), for example, found n-3 PUFA to decrease leptin gene expression in a dose- and time-dependent manner within a human trophoblast cell line. Alnajjar et al. (Reference Alnajjar, Chabane Sari and Abuharfeil19) described the effects of PUFA on the IL production on the basis of a dietary intervention study (Jordan population). A case–control study in the European Prospective Investigation into Cancer and Nutrition by Nieters et al. (Reference Nieters, Becker and Linseisen20) has obtained indications for possible interactive effects between dietary intake of PUFA and polymorphisms of different obesity candidate gene variants.
Obesity is the result of an interaction between genetic predisposition and the modern obesogenic environment(Reference Bouchard21). As it is spreading rather rapidly and presenting a major health problem, it is important to understand inter-individual variance and susceptibility. Thus, the aim of the present study is to investigate potential interactions between PUFA and selected SNP with respect to their impact on obesity risk.
Within the present study, twenty-one candidate genes for obesity, including cytokines, neurotransmitters, transcription factors and adipokines, were selected. Interaction effects with regard to obesity risk were calculated for the fatty acid composition of the erythrocyte membranes, a biomarker for dietary PUFA intake, and SNP located within these gene regions. The PUFA composition of the erythrocyte membranes reflects both PUFA intake and subsequent metabolism of the fatty acids over a period of weeks and months(Reference Arab and Akbar22). In order to lend more credence to the statistical results of the genetic IL-6–PUFA analyses, plasma IL-6 concentrations were also considered.
Methods
Study design and population
The second Bavarian Food Consumption Survey is a cross-sectional study, representative of the Bavarian population and designed to investigate dietary and lifestyle habits. German-speaking subjects (n 1050) aged 13–80 years were recruited between September 2002 and June 2003 following a three-step random route sampling procedure. A total of forty-two communities served as sampling points and were stratified by county and community characteristics. With a given start address, a random walk (every third household) was conducted and one random household member who met the inclusion criteria was selected. Information on the subjects' characteristics, lifestyle as well as health and socio-economic status were collected during a personal computer-assisted face-to-face interview at baseline. The participation rate was 71 %. A non-responder analysis was preformed. On average, non-responders had – among other characteristics – a higher BMI than the study participants. Within the following two weeks, data of the subjects' dietary intake and physical activity (PA) were assessed by three 24 h dietary recalls (two weekdays and one weekend day), which were conducted via telephone and by trained interviewers. For the 24 h dietary recalls, the software EPIC-Soft (International Agency for Research on Cancer, Lyon, France) was used(Reference Slimani, Deharveng and Charrondiere23–Reference Voss, Charrondiere and Slimani25). The participants had to recall their dietary intake as well as their PA of the previous day. All adult subjects ( ≥ 18 years) who completed at least one 24 h recall (n 879) were invited to their nearest public health office for blood sampling and standardised anthropometric measurements within 6 weeks after recruitment. Of these subjects, 65 % (n 568) accepted this invitation and represented the subgroup on which this evaluation is based on. The study was conducted according to the guidelines laid down in the Declaration of Helsinki, and all procedures involving human subjects were approved by the local ethics committee. Written informed consent was obtained from all study participants.
Anthropometric, dietary and physical activity assessment
Height was measured to the nearest 0·5 cm and weight to the closest 0·5 kg. BMI was calculated as weight/height2 (kg/m2). Subjects were classified according to the definition of the WHO(26) as obese (BMI ≥ 30 kg/m2) and non-obese (BMI < 30 kg/m2). Hip size was determined as the widest circumference measured over the buttocks and waist measurements were taken midway between the iliac crest and the margin of the lower rip.
The German food composition table BLS (Bundeslebensmittelschlüssel) (version II.3; BgVV, Berlin, Germany) was used to calculate nutrient intake. Data were weighted correspondingly to weekday or weekend day in order to calculate a mean daily intake per subject.
In the telephone interviews, participants were asked to recall their PA of the last 24 h. Standardised questions on type and duration of the PA in the categories of sports, occupation and other strenuous activities during leisure time as well as non-occupational television/personal computer use and duration of sleeping were part of the computer-based interview. Metabolic equivalents (MET) were matched to each activity and the energy expenditure (MET × h) of every individual was estimated(Reference Schaller, Seiler and Himmerich27).
Blood sampling
Venous blood was drawn, chilled at 4°C and further processed within 3 h. Plasma and buffy coat were separated from erythrocytes by centrifugation (2000 g for 15 min) before being divided into aliquots and stored at − 80°C for further analyses.
Fatty acid composition of erythrocyte membranes
Membrane fatty acid analysis was conducted using an aliquot of 0·5 ml erythrocyte suspension. After cell lyses through addition of aqua destillata, the erythrocyte membranes were isolated via centrifugation (20 000 g for 20 min at 4°C) and the pellet was resuspended with Tris-buffer (11 mm-Tris, 1 mm-Na-EDTA, pH 7·4); the washing procedure was repeated twice before adding 800 μl aqua destillata(Reference Golik, Weissgarten and Evans28). Fatty acid extraction was performed using a mixture of chloroform and methanol (2:1, v/v) according to a modification of the method described by Folch et al. (Reference Folch, Lees and Sloane Stanley29). The lipids were extracted twice using a chloroform–methanol mixture with the added antioxidant butylated hydroxytoluene (50 mg/l)(Reference Wren and Szczepanowksa30). The combination of these extracts was washed with a CaCl2 solution. The organic phase was collected and evaporated until dry. Resuspension of that extract was done using chloroform, and via transesterification with trimethylsulphonium hydroxide, the fatty acid methyl esters were obtained(Reference Butte31).
A 100 m CP-Sil-88 capillary column (Varian-Chrompack, Darmstadt, Germany), which was installed in an HP 5890 series II gas chromatograph with a flame-ionisation detector (Hewlett Packard, Munich, Germany), was used to identify and separate the different fatty acid methyl esters. Authentic standards (Sigma-Aldrich, Steinheim, Germany) were applied to assure a correct identification and quantification of the fatty acid methyl ester peaks. As a result, the content of twenty-two types of fatty acids was measured and is expressed as a percentage of the total fatty acid methyl esters (% fatty acid methyl esters) identified(Reference Hoff, Seiler and Heinrich32). For each sample, data represent the mean of two injections.
Analysis of plasma IL-6
Plasma IL-6 was measured by means of a commercial ELISA kit (Biosource, Brussels, Belgium). The intra- and inter-assay CV were below 7 and 9 %, respectively.
SNP selection
The genetic dataset was developed in 2006 and consists of different candidate genes, which were chosen on the basis of an extensive literature research. SNP covering these genes as well as 100 kb of region 5′ and 50 kb of region 3′ were selected based on hapmap data (www.hapmap.org; phases 1 and 2) with a minimum minor allele frequency of 0·05 according to the data of dbSNP Build 125 (an SNP database). For the present analyses, we carefully selected only genes for which an indication of a potential interaction with PUFA was provided by the literature. These include different cytokines and their receptors (IL-2, IL-6, IL-10, IL-18, TNF-α, TNFRSF1A, TNFRSF1B and TNFRSF21 (TNF receptor superfamily 1A, 1B and 21)), neurotransmitters and their receptors (NPY (neuropeptide Y), NPY1R, NPY5R (NPY receptors Y1 and Y5), MC4R (melanocortin 4 receptor), POMC (pro-opiomelanocortin), PPY (pancreatic polypeptide), and PYY (peptide YY)), transcription factors (PPARγ and PPARγC1A) and adipokines (LEP (leptin), LEPR (leptin receptor), ADIPOQ (adiponectin) and RETN (resistin)). For these genes, the genetic dataset holds a total number of 187 SNP, consisting of 180 tagging SNP, eighteen coding SNP and twelve candidate SNP previously reported to be associated with obesity. The median number of SNP per gene locus is 11 with a range of 2–16.
Genotyping and quality control
Genotyping was performed by GoldenGate Genotyping Assay (Illumina, Inc., San Diego, CA, USA) according to the standard protocol of the manufacturer. Additional inclusion criteria were a minor allele frequency of at least 5 % and genotyping call rate of not less than 95 %, leaving 157 SNP in the dataset.
Statistical analyses
The descriptive data are presented as median with 25 and 75 % quantiles for continuous parameters, as the majority of the variables were not normally distributed, or as percentage and absolute frequency for qualitative variables. Comparisons between the groups of obese and non-obese subjects were made by means of either the Mann–Whitney U test (continuous variables) or the Kruskal–Wallis test (qualitative parameters). Socio-economic status was categorised based on the values of three characteristics on a point scale including educational level, social position and the households' net income(Reference Winkler and Stolzenberg33). PA data represent the estimated overall energy expenditure (MET × h) of every individual(Reference Schaller, Seiler and Himmerich27).
Departure from the Hardy–Weinberg equilibrium was tested by means of an exact test(Reference Wigginton, Cutler and Abecasis34). All selected SNP were in the Hardy–Weinberg equilibrium except for rs1061624 (TNFRSF1B), rs16475 (NPY), rs16480 (NPY region 5′) and rs17366743 (ADIPOQ), applying a P value of 3·09 × 10− 4 (corrected for multiple testing) as the significance level.
For comparison of the allele frequencies between obese and non-obese subjects, a Kruskal–Wallis test was used. The main effects of SNP and PUFA as well as their interaction effects are derived from a logistic regression model assuming additive genetic effects, and are presented as OR with the corresponding 95 % CI. For the genetic main-effect models, SNP were introduced as discrete parameters with either three categories (homozygous wild type, heterozygous or homozygous mutant type) or two categories (homozygous wild type and one or more mutant allele carriers), depending on the number of subjects in the third category (homozygous mutant type; minimum of ten subjects). As EPA and DHA were highly correlated, their sum was used in the present analyses. The fatty acid variables (presented as percentage of fatty acid methyl esters) were established as continuous parameters. For the interaction models, both SNP and PUFA were introduced as continuous parameters. A likelihood ratio test was used to compare the models with and without an interaction term and a P value of < 0·05 was regarded as nominally statistically significant. These P values are denoted as P int. The interaction models were adjusted for sex, age, PA and socio-economic status and significant SNP–PUFA interactions were analysed further. The effect of each PUFA with respect to obesity risk was estimated within the gene strata (homozygous wild type and one or more mutant allele carriers) of the SNP using logistic regression models adjusted for sex, age, PA and socio-economic status. As the analyses were done within the SNP strata and the number of people carrying two minor alleles was often quite small, two categories were used for sample size reasons. We tried different procedures, inter alia by Bonferroni, Holm, Hochberg, Sidak or Benjamini and Hochberg, to correct the present results for multiple testing, but neither left us with any significant result. Therefore, all P values reported in the present study are uncorrected and only nominally significant at α 5 %. The SNP were tested for pairwise linkage disequilibrium (LD). If SNP were in high LD (r 2 ≥ 0·7), one of them was selected as representative for the LD block. We chose rs1800795 to represent the IL-6 LD block with rs1800797 (r 2 0·935) and rs2069833 (r 2 0·967) and IL-18 SNP rs3882891 to represent rs1946519 (r 2 0·77).
As five values of plasma IL-6 were declared as outliers (greater than mean plus five times the standard deviation), they were excluded from the analyses. While plasma IL-6 levels ranged between 0·25 and 11·64 pg/ml, the levels of those five outliers were substantially higher (51·5, 61·8, 68·92, 93·62 and 620·6 pg/ml). The reason for those high values could neither be clarified nor was a re-analysis of the samples possible. To account for skewness, the parameter was log-transformed. Plasma IL-6 is presented as geometric means and 95 % CI. All statistical analyses were performed with R software version 2.9.0 (R Development Core Team, 2009; http://www.r-project.org/).
Results
Table 1 summarises the characteristics of the study population. Obese subjects had a median BMI of 33·04 (25–75 % quantiles 31·19–36·12) kg/m2 and their median age exceeded that of the non-obese subjects (median BMI of 24·83 (25–75 % quantiles 22·55–27·20) kg/m2) by 12 years. The median hip and waist circumference of obese and non-obese participants differed by 12·5 and 22·5 cm and the plasma IL-6 level of obese subjects was also elevated. There was no significant difference in sex distribution, PA or dietary fatty acid intake between the two groups. The linoleic acid (LA), arachidonic acid (AA), and EPA+DHA compositions of erythrocyte membranes were higher for non-obese (v. obese) participants but only the difference in LA content reached statistical significance.
Table 1 Characteristics of obese (BMI ≥30 kg/m2) and non-obese (BMI <30 kg/m2) subjects in a subsample of the Bavarian Food Consumption Survey II
(Medians, 25–75 % quartiles, percentages or number of absolute frequencies)

MET, metabolic equivalents; % FAME in erythrocytes, fatty acids in percentage of total fatty acid methyl esters in erythrocyte membranes; % E, percentage of energy.
* Kruskal–Wallis test.
† Mann–Whitney U test.
‡ Values are geometric means and 95 % CI.
The following results are not corrected for multiple testing and therefore are only nominally significant at α 5 %. Table S1 of the supplementary material (available online at http://www.journals.cambridge.org/bjn) shows the distribution of the alleles within the two groups of obese and non-obese subjects, the P-values of the Kruskal–Wallis test and the main effects of all SNP analysed on the risk of obesity. Risk estimates were calculated for models with either three categories (homozygous wild type, heterozygous and homozygous mutant type) or two categories (homozygous wild type and one or more mutant allele carriers). The P value of the continuous model is also given as a P trend. A nominal P value < 0·05 was reached by rs4719714 and rs12700386 (IL-6 region 5′), rs2069849 (IL-6), rs1061628 (TNFRSF1B) and rs1116656 (LEP region 3′). The crude main effects for LA (OR 0·90, 95 % CI 0·83, 0·98), AA (OR 0·97, 95 % CI 0·93, 1·01) and EPA+DHA (OR 0·96, 95 % CI 0·89, 1·03) in erythrocytes showed an indication of an inverse association with obesity, but only for LA, the statistical significance was reached.
SNP for which the interaction term with PUFA reached nominal statistical significance (α 5 %) were stratified by genotype (two categories: homozygote wild type and one or more mutant allele carriers), and risk estimates were calculated for the corresponding PUFA within these strata (Tables 2–4) .
Table 2 Significant SNP–linoleic acid interactions on obesity risk, showing the adjusted* relative risk of obesity per 1 mol% increase of linoleic acid† in erythrocyte membranes by allelic variants
(Odds ratios and 95 % confidence intervals)

* Adjusted for age, sex, physical activity and socio-economic status.
† Main effect: OR 0·90, 95 % CI 0·82, 0·99.
‡ P value of the likelihood-ratio test (adjusted continuous interaction model).
Table 3 Significant SNP–arachidonic acid interactions on obesity risk, showing the adjusted* relative risk of obesity per 1 mol% increase of arachidonic acid† in erythrocyte membranes by allelic variants
(Odds ratios and 95 % confidence intervals)
* Adjusted for age, sex, physical activity and socio-economic status.
† Main effect: OR 0·97, 95 % CI 0·94, 1·01.
‡ P value of the likelihood-ratio test (adjusted continuous interaction model).
Table 4 Significant SNP–EPA+DHA interactions on obesity risk, showing the adjusted* relative risk of obesity per 1 mol% increase of EPA+DHA† in erythrocyte membranes by allelic variants
(Odds ratios and 95 % confidence intervals)

* Adjusted for age, sex, physical activity and socio-economic status.
† Main effect: OR 0·95, 95 % CI 0·89, 1·02.
‡ P value of the likelihood-ratio test (adjusted continuous interaction model).
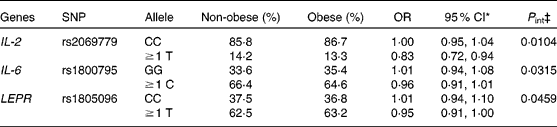
Concerning cytokine genes, we found several SNP–PUFA interactions in relation to obesity risk. For IL-2, the interaction term between rs2069779 and all three PUFA as well as between rs2069762, rs4833248 (IL-2 region 5′) and LA were statistically significant. Each PUFA associated with rs2069779 had a decreasing effect on obesity risk for minor allele carriers (Fig. 1). In this gene stratum, the risk decreased for each mol % increase in erythrocyte membrane-bound PUFA with OR of 0·63 (LA), 0·83 (AA) and 0·66 (EPA+DHA). For rs4833248 as well as rs2069762, homozygous wild-type carriers benefited from increased LA content in erythrocyte membranes. Analyses for IL-6 revealed three SNP: rs1800795; rs10242595; rs2069861. The first two showed a significant interaction effect with LA with P int = 0·0341 and 0·0315, respectively. For each percentage increase in erythrocyte membrane-bound LA, obesity risk decreased in carriers of at least one minor allele of rs1800795 with an OR of 0·86 (95 % CI 0·76, 0·96); a similar effect was found for the rs1800795–AA interaction (Fig. 2). In the case of rs10242595, the relative risk decreased for carriers of the homozygous wild-type alleles with increasing LA content (OR 0·81, 95 % CI 0·71, 0·93). The interaction effect of rs2069861 and EPA+DHA was significant as well. Here, the obesity risk decreased with each mol % increase of erythrocyte membrane-bound EPA+DHA for minor allele carriers (OR 0·74, 95 % CI 0·57, 0·94). For IL-18, one SNP (rs3882891) interacted significantly with LA when analysing obesity risk. Minor allele carriers of this SNP had a reduced obesity risk with increasing LA content in erythrocyte membranes (OR 0·84, 95 % CI 0·75, 0·94, P int = 0·0203).
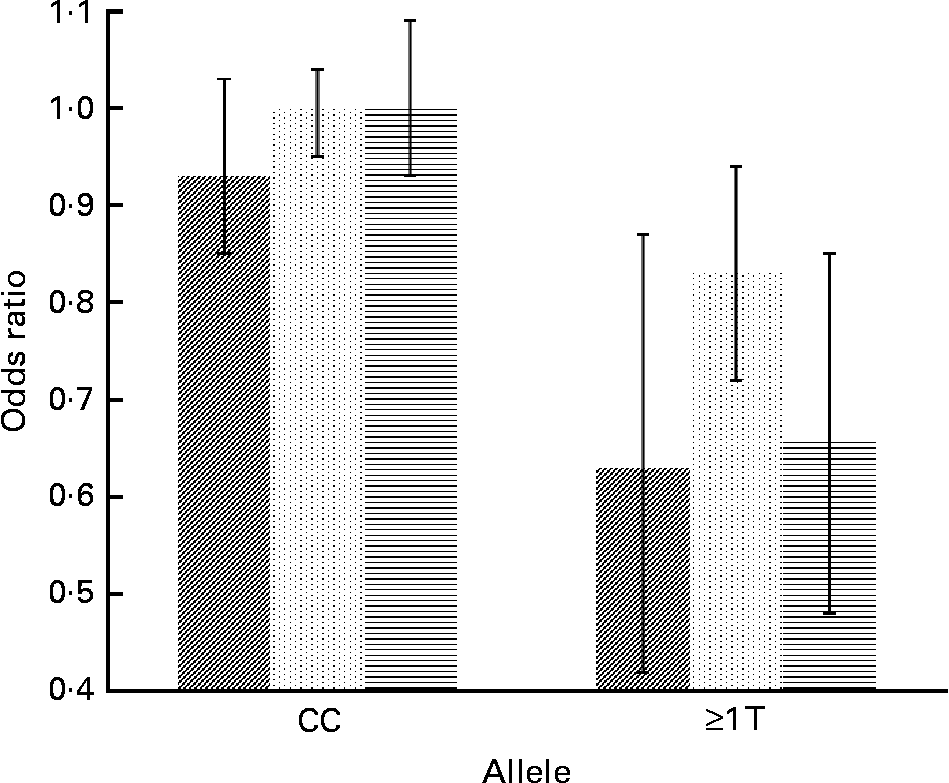
Fig. 1 Effect of linoleic acid (![]() , P int = 0·0310), arachidonic acid (
, P int = 0·0310), arachidonic acid (![]() , P int = 0·0104) and EPA+DHA (
, P int = 0·0104) and EPA+DHA (![]() , P int = 0·0022) on the risk of obesity, stratified by genotype of IL-2 (rs2069779). Estimates are adjusted for age, sex, physical activity and socio-economic status. P int, P value of the likelihood-ratio test comparing models with and without an interaction term.
, P int = 0·0022) on the risk of obesity, stratified by genotype of IL-2 (rs2069779). Estimates are adjusted for age, sex, physical activity and socio-economic status. P int, P value of the likelihood-ratio test comparing models with and without an interaction term.
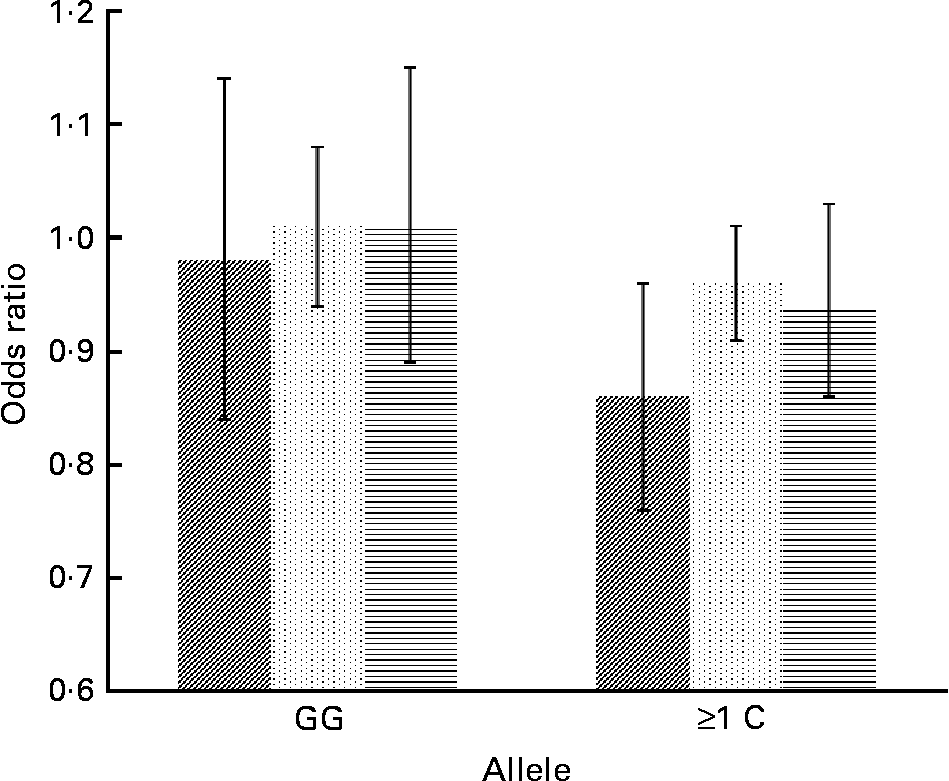
Fig. 2 Effect of linoleic acid (![]() , P int = 0·0341), arachidonic acid (
, P int = 0·0341), arachidonic acid (![]() , P int = 0·0315) and EPA+DHA (
, P int = 0·0315) and EPA+DHA (![]() , P int = 0·0878) on the risk of obesity, stratified by genotype of IL-6 (rs1800795). Estimates are adjusted for age, sex, physical activity and socio-economic status. P int, P value of the likelihood-ratio test comparing models with and without an interaction term.
, P int = 0·0878) on the risk of obesity, stratified by genotype of IL-6 (rs1800795). Estimates are adjusted for age, sex, physical activity and socio-economic status. P int, P value of the likelihood-ratio test comparing models with and without an interaction term.
We also obtained evidence for SNP–EPA+DHA interaction effects of the TNFRSF1B gene. Subjects carrying at least one minor allele of rs3766730 (OR 0·86, 95 % CI 0·75, 0·98, P int = 0·0225) or rs2275416 (OR 0·88, 95 % CI 0·78, 0·99, P int = 0·0455) had a lower obesity risk than homozygous wild-type carriers. In relation to obesity risk, two SNP of TNFRSF21 were shown to interact with EPA+DHA; rs9381530 (region 3′) with an OR of 0·79 (95 % CI 0·68, 0·91, P int = 0·0065) and rs2236039 with an OR of 0·87 (95 % CI 0·78, 0·96, P int = 0·0482). In each case, obesity risk decreased with increasing PUFA content in subjects carrying two major alleles.
Among the selected adipokine genes, few significant results were obtained. The interaction terms of two ADIPOQ SNP (rs1063539 and rs2241766) and membrane EPA+DHA content were significantly associated with obesity risk. In homozygous wild-type carriers, obesity risk decreased with increasing EPA+DHA content (Table 4). Concerning a leptin receptor gene polymorphism (rs1805096), an inverse association with obesity risk existed in carriers of the minor allele, with an increasing erythrocyte membrane content of LA (OR 0·83, 95 % CI 0·74, 0·93, P int = 0·0084; Table 2) or AA (OR 0·95, 95 % CI 0·91, 1·00, P int = 0·0459; Table 3).
Table 5 gives the geometric means and the 95 % CI of plasma IL-6 stratified by IL-6 SNP rs1800795, rs10242595 and rs2069861 (homozygote wild type and one or more mutant alleles) and tertiles of LA, AA and EPA+DHA for obese and non-obese subjects. The plasma IL-6 concentrations were generally higher in obese subjects compared with non-obese subjects (see also Table 1) and decreased with increasing PUFA content in erythrocyte membranes (tertiles). These results lend credit to the identified IL-6 SNP–PUFA interactions and obesity risk derived from the statistical models.
Table 5 Plasma IL-6 concentrations (pg/ml) by tertiles of linoleic acid, arachidonic acid and EPA+DHA in erythrocyte membranes by allelic variants of IL-6 SNP, rs1800795, rs10242595 and rs2069861
(Geometric mean values and 95 % confidence intervals)

Discussion
The present study aimed to investigate the additional effect on obesity brought on by the interaction of selected genetic variants and PUFA content of erythrocyte membranes. Out of the four different groups of genes, including cytokines, adipokines, neurotransmitters and transcription factors, we obtained significant interaction effects between the SNP of IL-2, IL-6, IL-18, TNFRSF1B, TNFRSF21, LEPR or ADIPOQ and PUFA content in erythrocyte membranes. We found a reduced obesity risk for minor allele carriers of most variants with high PUFA content in erythrocyte membranes, except for the SNP of TNFRSF21, ADIPOQ, rs2069762 (IL-2), rs4833248 (IL-2 region 5′) and rs10242595 (IL-6 region 3′). With the latter genes, subjects, homozygote for the major allele, benefited from an increased PUFA content of erythrocyte membranes. In the case of IL-6, the analysed plasma IL-6 protein concentration supports the statistical findings.
Obesity has been described as a state of chronic low-grade inflammation(Reference Engstrom, Hedblad and Stavenow35, Reference Festa, D'Agostino and Williams36). Thus, polymorphisms in different cytokines were included in the analyses of gene–PUFA interactions. Cytokines are a group of modulatory proteins which respond to various stimuli, thereby activating second messengers and signal transduction pathways within the cells(Reference Smith and Humphries37). Several cell studies and also dietary intervention studies have shown a reduced production of IL-2 in response to PUFA(Reference Merzouk, Saker and Reguig16, Reference Verlengia, Gorjao and Kanunfre18, Reference Alnajjar, Chabane Sari and Abuharfeil19, Reference von Schacky38); however, the exact mechanism behind this phenomenon still remains unclear(Reference Calder and Grimble39, Reference Gorjao, Hirabara and de Lima40). In the present study, obesity risk decreased with increasing PUFA content of the erythrocyte membranes, and thus confirms the expected direction. PUFA eicosanoid derivates, such as PGE2, are involved in the modulation of the intensity and duration of inflammatory processes and suppress the production of IL-6(Reference Stulnig10). Adipose tissue in human subjects releases IL-6 and serum levels are positively correlated with body fat mass(Reference Vozarova, Weyer and Hanson41). Himmerich et al. (Reference Himmerich, Fulda and Linseisen42) confirmed this relation for the present population. IL-6 gene transcription was found to be influenced in vitro by the rs1800795 polymorphism within the promoter region(Reference Fishman, Faulds and Jeffery43). The G allele of this SNP was described as to be more common in lean subjects(Reference Berthier, Paradis and Tchernof44); additionally, a lower BMR was measured in subjects with the CC genotype(Reference Kubaszek, Pihlajamaki and Punnonen45), which might eventually predispose to weight gain; this hypothesis has been supported by some studies but not confirmed in two meta-analyses of the association of this SNP with BMI(Reference Huth, Illig and Herder46, Reference Qi, Zhang and van Dam47). Our findings on SNP–PUFA interactions may provide an explanation for the diverging results since they consider the possible interplay between the SNP and PUFA supply status. For the other IL-6 SNP, rs10242595, the A variant was found to be significantly associated with decreased fat mass in young adult men, a result which was replicated in two other population-based studies of elderly men(Reference Andersson, Strandberg and Nilsson48). The interaction effect between this SNP and PUFA in the present study is indicative towards the importance of this SNP for the development of obesity. Variations in the IL-18 gene have been associated with IL-18 plasma concentrations and measures of obesity(Reference Thompson, Sanders and Stephens49). Obese subjects show higher levels of IL-18 than lean subjects(Reference Skurk, Kolb and Muller-Scholze50), and IL-18 has been associated with excess adiposity(Reference Hung, McQuillan and Chapman51). Our IL-18 variant is in complete LD with rs5744292, an IL-18 SNP whose minor allele has been reported to be associated with lower circulating IL-18 levels and lower mRNA expression in immortalised lymphocytes(Reference Barbaux, Poirier and Godefroy52, Reference Tiret, Godefroy and Lubos53). Furthermore, a suppressing effect of PGE2 on the expression of IL-18 has been shown in cell studies(Reference Suk, Yeou Kim and Kim54). The finding of an IL-18–PUFA interaction in the present study fits well with these data. Even though the production of TNF-α by monocytes and macrophages is also suppressed through PGE2(Reference Stulnig10), no significant interactions could be determined within the present study. However, we found evidence for significant interaction effects for its receptors, TNFRSF1B and TNFRSF21.
Overall, all identified (significant) interactions between cytokine SNP and PUFA indicate an inverse association with obesity risk for minor allele carriers, with increasing PUFA content in erythrocyte membranes, except for SNP of TNFRSF21, rs4833248 (IL-2 region 5′) and rs10242595 (IL-6 region 3′).
Besides its role for lipid storage, adipose tissue functions as an endocrine organ, regulating metabolism and different vital functions related, among others, to inflammation(Reference Saltiel55, Reference Spiegelman and Flier56). Thus, different adipokines have been included in the present analyses. Adiponectin is exclusively secreted by adipose tissue and serum levels are inversely correlated with body fat mass(Reference Arita, Kihara and Ouchi57). The mRNA expression is reduced in obese individuals(Reference Hu, Liang and Spiegelman58). Serum adiponectin levels are highly heritable (approximately 50 %) and are linked to the ADIPOQ gene locus(Reference Chuang, Chiu and Sheu59–Reference Vasseur, Helbecque and Dina61). Different cell and dietary intervention studies found EPA+DHA to stimulate the expression of ADIPOQ and to increase plasma adiponectin levels(Reference Itoh, Suganami and Satoh62–Reference Yu, Lin and Wu64); however, findings differ(Reference Lorente-Cebrian, Perez-Matute and Martinez65). EPA+DHA might possibly up-regulate ADIPOQ by acting through PPARγ, affecting the ADIPOQ promoter(Reference Iwaki, Matsuda and Maeda15). The present results are in line with these findings and show a significantly decreased obesity risk for carriers of two major alleles of rs2241766 or rs1063539, with increasing EPA+DHA concentrations in erythrocyte membranes. Different animal, human and cell studies have shown an inverse effect of PUFA on the LEP mRNA expression(Reference Reseland, Haugen and Hollung17, Reference Phillips, Goumidi and Bertrais66). The present analyses resulted in one significant interaction for a variant of the leptin receptor gene, which is in line with these findings.
We see two major mechanisms of how PUFA may in conjunction with genetic variants affect obesity risk: either via direct modification of gene transcription or by products of the eicosanoid pathway. To compare the direction of the different effects of PUFA on the risk of obesity within the SNP strata of the significant interaction models, we also estimated the effects of the remaining PUFA by the given SNP strata for which the interaction term with those SNP was not significant (e.g. Fig. 2). We observed quite similar effects (direction and estimates) over the different PUFA in the various SNP strata (data not shown). A high erythrocyte membrane content of LA or AA or EPA+DHA thereby did either show no association or an inverse association with obesity risk in each of the SNP strata. Therefore, we conclude that PUFA exert their effects rather via modification of gene transcription than through metabolites derived during eicosanoid synthesis, since the latter would have led to differential effects of n-3 and n-6 PUFA. In mutually adjusted analyses, we have also not received any indication for changes of effects of n-3 PUFA-adjusted n-6 PUFA and vice versa. It is important to mention that the frequency of these SNP for which we observed interactive effects with PUFA is fairly high, except for IL-2 SNP (rs2069779). This implicates that a substantial part of the population would benefit from a high PUFA intake with respect to obesity risk.
A major limitation of the present study is obviously the small sample size. Studies of genetic associations with complex diseases need thousands of cases and controls(Reference Colhoun, McKeigue and Davey Smith67); however, this requirement is not easily fulfilled with respect to the costly fatty acid analyses. The small number of cases and controls and consequently the limited statistical power strongly argue for a careful interpretation of the results and a replication in a second, larger and independent study. Because of the small sample size and the resulting limited power, we did not correct for multiple testing. No correction method for multiple testing left us with any significant result. Therefore, all reported P values are not corrected and are only nominally significant at α 5 %. The observational nature of the study does not allow for interpreting causal associations, and we cannot rule out the possibility of reverse causation. However, we controlled for potential confounding by adjusting for sex, age, PA and socio-economic status.
The major strength of the present study is the use of erythrocyte membranes to assess biologically available PUFA at the cellular level and its association with genetic variants influencing the risk of obesity. With the utilisation of biomarkers as an objective metabolic correlate of dietary PUFA intake, misclassifications can be largely avoided. A further strength of the present study is its population-based design aiming at representativeness for the adult Bavarian population and the strict quality control in the analyses.
Acknowledgements
The authors acknowledge the cooperation of all study participants. We thank Georg Karg, Kurt Gedrich and Stefanie Himmerich for their major contribution in the set-up and conduct of the study. The study was supported by funds of the Bavarian Ministry of Environment, Health and Consumer Protection and the Kurt-Eberhard-Bode-Stiftung. The authors' contributions were as follows: C. J. performed the statistical analyses and drafted the manuscript. S. K., A. N., H. H. and M. A. K. were responsible for SNP selection and genotyping; S. K. also wrote these sections of the manuscript. H. S. contributed to the data collection. H.-E. W., G. W. and S. L. provided critical revision. C. G. gave statistical advice. J. L. was responsible for the study design, fatty acid data acquisition as well as supervision and together with G. W. for funding. All authors contributed to the interpretation and discussion of the results and read and approved the final version of the manuscript. The present study represents original work that has not been published previously and the authors declare that there are no conflicting interests.









