Dietary intake of n-3 long-chain PUFA (LCPUFA) modulates the plasma lipid profile in adults mainly by lowering circulating TAG, and sometimes also increasing LDL- and HDL-cholesterol(Reference Harris, Miller and Tighe1). We have also demonstrated this in a few studies in children(Reference Harslof, Damsgaard and Hellgren2,Reference Pedersen, Molgaard and Hellgren3) . However, results from intervention studies vary, and there is limited knowledge about the impact of genotype on the response to n-3 LCPUFA(Reference Minihane4), especially in children. Such insight may help understand the mechanisms of action of n-3 LCPUFA and identify persons who may benefit the most from an increased consumption.
Fish is the main source of the n-3 LCPUFA, EPA and DHA, but n-3 LCPUFA are also endogenously produced from α-linolenic acid by the action of the fatty acid desaturase (FADS) enzymes. SNP in the FADS1 and FADS2 genes, which encode the D5- and D6-desaturase, respectively, have been shown to reduce enzyme activities and endogenous LCPUFA production and status(Reference Lauritzen, Sorensen and Harslof5–Reference Marquardt, Stohr and White8). The rs1535 SNP in the intronic region of FADS2 and rs174448 between FADS2 and FADS3, which tags SNP in FADS2 genes, has both been associated with n-3 LCPUFA status in Danish infants(Reference Harslof, Larsen and Ritz6). Thus, FADS SNP may help explore the role of n-3 LCPUFA status on health outcomes. Carriage of FADS minor alleles has been linked to higher plasma TAG and lower HDL- and LDL-cholesterol in adults(Reference Kathiresan, Willer and Peloso9,Reference Aulchenko, Ripatti and Lindqvist10) , and comparable associations have been observed in a few studies in children and infants(Reference Standl, Lattka and Stach11,Reference Lauritzen, Amundsen and Damsgaard12) .
The effects of n-3 LCPUFA on the plasma lipid profile may involve changes in the efficiency of lipid transport and apoE-mediated lipid uptake in the tissues(Reference Dallongeville, Lussier-Cacan and Davignon13). The APOE4 variant of the gene that encodes apoE has been associated with higher plasma LDL-cholesterol and risk of CHD, whereas the rare APOE2 variant has been shown to reduce receptor binding and to be associated with lower LDL-cholesterol(Reference Song, Stampfer and Liu14,Reference Minihane, Jofre-Monseny and Olano-Martin15) . However, few studies have examined the responsiveness of carriers of the APOE variants to n-3 LCPUFA intervention(Reference Minihane4).
Experiments in rodents show that n-3 LCPUFA intake can reduce plasma insulin and glucose(Reference Perez-Matute, Perez-Echarri and Martinez16–Reference Storlien, Kraegen and Chisholm19), whereas randomised trials in humans show no or inconsistent effects on glucose homoeostasis markers(Reference Damsgaard, Frokiaer and Andersen20–Reference Pelikanova, Kohout and Valek24). The inconsistencies may be due to genotypic differences, since emerging evidence suggest that the effects of n-3 LCPUFA depend on polymorphisms in genes that encode proteins involved in the metabolic pathways or mechanisms of action of these fatty acids(Reference Minihane4). We recently found that fish oil supplementation only reduced plasma glucose in infants who were minor allele carriers of the rs1801282 SNP in the PPAR-γ2 gene PPARG2 (Reference Harslof, Damsgaard and Hellgren2). PPAR-γ2 is mainly expressed in adipose tissue where it regulates adipogenesis, lipid metabolism and insulin sensitivity(Reference Tontonoz and Spiegelman25), whereas PPAR-α is a major regulator of lipid metabolism in the liver(Reference Tai, Corella and Demissie26) and both n-3 LCPUFA and eicosanoids are ligands for these transcription factors. The minor allele of rs1800206 in the PPARA gene has been shown to increase LDL-cholesterol(Reference Maciejewska-Skrendo, Buryta and Czarny27) and to modify associations between PUFA intake and the lipid profile in adults(Reference Tai, Corella and Demissie26,Reference Chan, Tan and Deurenberg-Yap28) . However, its effect on gene transcription may depend on the concentration of ligand(Reference Maciejewska-Skrendo, Buryta and Czarny27).
In a cross-sectional study among 713 Danish 8–11-year-old children participating in the OPUS School Meal Study, we previously showed that whole-blood EPA was inversely associated with plasma TAG and positively with LDL- and HDL-cholesterol, whereas DHA was inversely associated with serum insulin(Reference Damsgaard, Eidner and Stark29). In the same cohort, we recently found that FADS minor alleles modulated the children’s LCPUFA status, that is, minor allele carriers of rs174448 had lower n-3 LCPUFA and arachidonic acid in whole blood, whereas carriers of rs1535 minor allele mainly had lower arachidonic acid(Reference Lauritzen, Sorensen and Harslof5). In the present study, we aimed to explore (1) whether FADS polymorphisms were associated with TAG, HDL- and LDL-cholesterol, insulin and glucose in order to get insight into the role of these genotypes in children’s cardiometabolic health and (2) whether polymorphisms in PPAR genes and APOE modified associations between FADS or n-3 LCPUFA and the cardiometabolic blood markers, as outlined in Fig. 1. This may help us understand the extent to which PPAR and apoE mediate the effects of n-3 LCPUFA and if some children benefit more from a given n-3 LCPUFA status than others.
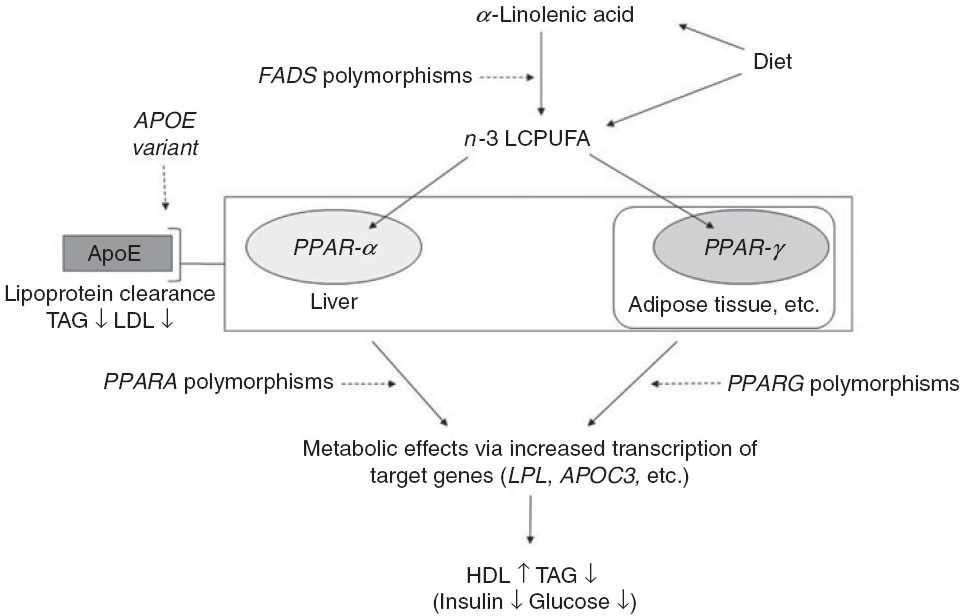
Fig. 1. Outline of the idea of the paper. Conversion of dietary α-linolenic acid to n-3 long-chain PUFA (LCPUFA) involves the desaturase enzymes encoded by the fatty acid desaturase (FADS) genes and the enzyme’s activity (and thereby the conversion) are affected by SNP in FADS. n-3 LCPUFA may also be consumed preformed in fish and fish oils and are ligands for transcription factors such as PPAR-α and PPAR-γ. These are found mainly in the liver and in adipose tissue, respectively, and are involved in the effects of n-3 LCPUFA on circulating HDL-cholesterol, TAG and markers of glucose homoeostasis. SNP in their genes PPARA and PPARG affect transcription of target genes thereby probably modulating the effects of n-3 LCPUFA. Genetic variants of the apoE gene APOE affect lipoprotein clearance, but the effects in combination with n-3 LCPUFA are not clear.
Methods
Study design and participants
The study was based on cross-sectional baseline data from the Optimal well-being, development and health for Danish children through a healthy New Nordic Diet (OPUS) School Meal Study, which investigated the effects of Nordic school meals on children’s cardiometabolic health, well-being and cognitive performance(Reference Damsgaard, Dalskov and Petersen30). It was conducted according to the guidelines in the Declaration of Helsinki, approved by the Danish National Committee on Biomedical Research Ethics (no. H-1-2010-124), and the baseline study was registered at clinicaltrials.gov as NCT01577277. All children from third and fourth grade at nine schools in different socio-economic areas in the Eastern part of Denmark were invited to participate in the study, and baseline assessments were performed from August to December 2011. Schools were invited by telephone and included if they had at least four classes at the third and fourth grade level, suitable kitchen facilities available for school meal production and high motivation for participation as determined by the study team(Reference Damsgaard, Dalskov and Petersen30). Children were excluded only if they had severe food-related allergies, food intolerances, or malabsorption, severe mental handicaps or were participating in other research projects that involved blood sampling or radiation. The original power calculation for the OPUS School Meal Study showed that 673 completing children were needed in order to detect a 0·32 point intervention difference in the primary outcome, a metabolic syndrome score(Reference Damsgaard, Dalskov and Laursen31). A total of 1021 children were invited of which parents of 834 children (82 %) gave informed written consent to their child’s participation(Reference Damsgaard, Dalskov and Petersen30). The current paper is based on data from the 757 children who had available data on at least one of the investigated SNP and at least one of the cardiometabolic markers.
Sociodemographics and pubertal status
Parental education was evaluated as the highest education level in the household, based on interview with the parents, and children’s ethnicity was characterised as Caucasian, Asian, African American or Latin American by the investigators. Pubertal status was self-evaluated by the child with parental assistance if necessary, using five categories (Tanner stages) of breast development in girls and pubic hair in boys(Reference Morris and Udry32).
Blood sampling and anthropometry
Blood sampling and anthropometry were performed by standard procedures in the morning in an air-conditioned double-decker truck equipped as a mobile laboratory. All children reported to have fasted overnight except for twenty-two children of whom nine had only consumed chewing gum or single bites of food. Local anaesthetic patches (EMLA; Astra Zeneca) were provided, and venous blood was drawn from the antecubital vein. Height was measured three times to the nearest 0·1 cm using a portable stadiometer (CMS Weighing Equipment), with the children holding their heads in the Frankfurt horizontal plane, and the mean was calculated. Body weight was measured to the nearest 0·1 kg on a digital scale (Tanita 800S; Tanita) with the child in underwear and without shoes. Sex- and age-adjusted z-scores for BMI were calculated using WHO AnthroPlus software(33), and the prevalence of underweight, overweight and obesity was determined based on age- and sex-specific cut-offs as described by Cole et al.(Reference Cole, Bellizzi and Flegal34,Reference Cole, Flegal and Nicholls35) .
Blood analyses
Glucose was assessed in fresh blood by a Hemocue Glucose 201 (Hemocue Denmark) calibrated to calculate plasma concentrations from whole blood. Blood for insulin analysis was collected in serum tubes with gel and left to coagulate for 30 min at room temperature. Serum and heparinised plasma for measurement of cholesterols and TAG were obtained by centrifugation at 2500 g for 10 min. Heparinised whole blood was mixed with 0·1 % butylated hydroxytoluene (Sigma-Aldrich) in ethanol (0·1 ml per ml blood) for fatty acid composition determinations. All blood fractions were stored at –80°C until analysis.
Serum insulin was measured by an automated chemiluminescent immunoassay on an ADVIA Centaur XP (Siemens Healthcare). Plasma total and HDL-cholesterol and TAG were measured on a Vitros 5.1 FS (Ortho-Clinical Diagnostics). LDL-cholesterol concentrations were calculated by Friedewald’s equation(Reference Friedewald, Levy and Fredrickson36). The inter- and intra-assay CV were 1·4 and 1·2 % (total cholesterol); 2·0 and 1·2 % (HDL-cholesterol); 1·5 and 0·8 % (TAG); and 2·5 and 3·1 % (insulin).
Whole-blood fatty acid composition was measured by high-throughput GC within 3 months after blood sampling as previously described(Reference Damsgaard, Eidner and Stark29,Reference Armstrong, Metherel and Stark37,Reference Metherel, Buzikievich and Charkhzarin38) . Briefly, thirty-two individual fatty acids were determined quantitatively by comparison with internal (22 : 3n-3 ethyl ester; Nu-Check Prep) and external reference standards (GLC-462, Nu-Check Prep). Lipids were extracted using chloroform:methanol (2:1, v/v) and transesterified to fatty acid methyl esters using 14 % boron trifluoride in methanol prior to GC analyses with flame ionisation detection using a Varian 3900 gas chromatograph equipped with a DB-FFAP 15 m × 0·10 mm injected dose × 0·10 μm film thickness capillary column (J&W Scientific from Agilent Technologies). n-3 LCPUFA were calculated as EPA + DHA in % of the total fatty acids. The intra- and inter-assay CV were 1·3 and 4·5 % for EPA and 2·4 and 6·4 % for DHA, respectively.
Genotyping and selection of SNP
Buffy coat DNA extraction and genotyping were performed at LGC Genomics Ltd using their Competitive Allele Specific Polymerase Chain Reaction genotyping technology. We included six functional SNP that have been associated with either blood LCPUFA, lipid profile or glucose metabolism and/or were regulated by LCPUFA in previous studies(Reference Harslof, Damsgaard and Hellgren2,Reference Harslof, Larsen and Ritz6,Reference Dallongeville, Lussier-Cacan and Davignon13,Reference Tai, Corella and Demissie26,Reference Chan, Tan and Deurenberg-Yap28) : FADS rs1535 and rs174448, PPARA rs1800206, PPARG2 rs1801282, as well as APOE rs7412+rs429358 which combined determine APOE genotype(Reference Minihane4) (online Supplementary Tables S1 and S2).
Statistical analysis
Data were analysed with SPSS version 23 (IBM Corporation) and R version 3.5.1 (The R Foundation for Statistical Computing Platform), and statistical significance was established at P < 0·05. Hardy–Weinberg equilibrium was assessed by χ 2 test(Reference Rodriguez, Gaunt and Day39) and linkage disequilibrium by use of the R package ‘genetics’. Characteristics of included and non-included children were compared using unpaired t test (age and BMI z-score) or χ 2 test (sex and parental education), and allele frequencies were compared between ethnic groups by χ 2 test. Cardiometabolic blood markers (plasma TAG, HDL-cholesterol, LDL-cholesterol and glucose and serum insulin) were checked for normal distribution by visual inspection of qq-plots and histograms, and TAG and insulin were ln-transformed to obtain normality. For each SNP, children were categorised as major allele homozygous (MM) or minor allele carriers (Mm+mm). We generated APOE genotypes based on the following rs7412/rs429358 allele combinations: mm/MM (E2/E2), Mm/MM (E3/E2), MM/MM (E3/E3), MM/Mm (E3/E4), MM/mm (E4/E4) and Mm/Mm (E2/E4) (online Supplementary Table S2). None of the children was carriers of E1. In order to assess the effects of E2 and E4 carriage compared with the more common E3/E3 genotype, we excluded twenty-one children with E2/E4 and pooled E3/E2 + E2/E2 as well as E3/E4 + E4/E4. Cardiometabolic characteristics of the APOE genotypes were compared by one-way ANOVA.
To test if the FADS SNP were associated with the cardiometabolic outcomes, we fitted linear mixed models including school and class as random effects, and age, height and BMI z-score as fixed effects or covariates, in order to reduce variation. Puberty and sex were included in the models as a sex × puberty interaction term, due to the different puberty scales used for boys and girls. Linear mixed models were fitted to test whether the PPAR SNP (rs1801282 or rs1800206) or APOE genotype showed interaction with the FADS SNP (rs1535 or rs174448), or with whole-blood n-3 LCPUFA. For all of the outcomes, these models included one interaction term at a time as well as the same adjustments as described above. Each statistical model included only participants with available data on the specific SNP and outcome. To check for consistency, all analyses were also conducted without any adjustments, in secondary analyses.
Results
Children’s characteristics
There was an even distribution of boys and girls, most children were of Caucasian origin and had normal weight, and about 1/3 (mainly girls) had entered puberty (Table 1). The 757 included children comprised 91 % of the original OPUS School Meal Study population and did not differ from the non-included children with regard to age, sex, BMI z-scores or parental education (P > 0·24, data not shown). The minor allele frequencies of the six investigated SNP ranged from 6 to 36 % and showed no deviations from Hardy–Weinberg Equilibrium (online Supplementary Table S1). As expected, the two FADS SNP (rs1535 and rs174448) were somewhat in linkage disequilibrium (D = 0·015, r 2 0·43), but all other SNP combinations showed little indication of linkage (D between –0·001 and 0·008, r 2 < 0·006). There were no differences in allele frequencies between ethnic groups for any of the SNP (P > 0·19).
Table 1. Baseline characteristics of the 757 children in the study
(Mean values and standard deviations; median values and interquartile ranges (IQR); percentages)
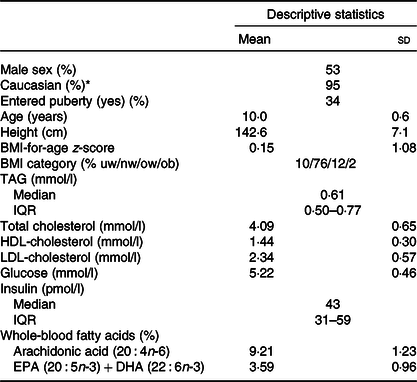
uw, Underweight; nw, normal weight; ow, overweight; ob, obese.
* The remaining children were of Asian (2 %), African American (2 %) and Latin American (1 %) origin.
Associations between FADS genotypes and the cardiometabolic markers
Major allele homozygotes of rs174448 had 0·04 (95 % CI 0·00, 0·07) mmol/l lower plasma TAG and 0·04 (95 % CI 0·00, 0·09) mmol/l higher HDL-cholesterol than minor allele carriers (Fig. 2). Comparable effect sizes in the same direction were seen for rs1535, but these did not reach statistical significance (online Supplementary Table S3). In addition, minor allele carriers of both rs174448 and rs1535 tended to have lower plasma glucose (P < 0·10, online Supplementary Table S3). There were no other differences, and unadjusted analyses showed the same results (data not shown).
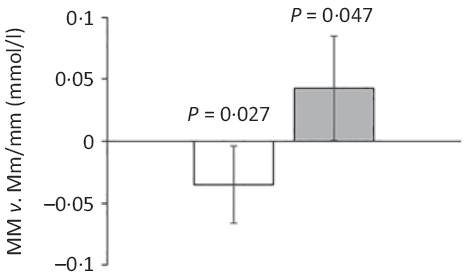
Fig. 2. Estimated differences in TAG and HDL-cholesterol between major allele homozygotes and minor allele carriers of the fatty acid desaturase (FADS) rs174448 genotype. Bars and error lines indicate estimated differences and 95 % confidence intervals, respectively, for MM compared with Mm/mm adjusted for age, sex × puberty, height, BMI z-score, school and class (n 316 for MM and n 440 for Mm/mm). P values are shown for these comparisons. ![]() , TAG;
, TAG; ![]() , HDL.
, HDL.
Effect modification by PPAR SNP
None of the associations between FADS and the cardiometabolic markers was modified by rs1800206 (PPARA) or rs1801282 (PPARG2) (all P interaction > 0·10, data not shown). Furthermore, there were no interaction between n-3 LCPUFA and the PPARA SNP (online Supplementary Table S4). However, as shown in Table 2, the associations between n-3 LCPUFA and TAG, LDL-cholesterol and insulin were modified by PPARG2 genotype. The associations in minor allele carriers were inverse, whereas there were no or positive associations in major allele homozygotes (Table 2). There was no interaction for HDL-cholesterol or glucose (Table 2). Repeating the analyses without adjustments did not change the results (data not shown).
Table 2. Associations between n-3 long-chain PUFA (LCPUFA) and the cardiometabolic markers according to PPARG2 rs1801282 genotype*
(β Coefficients and 95 % confidence intervals)
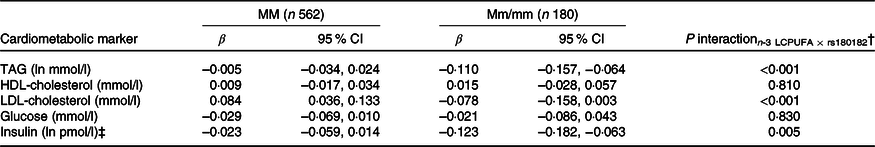
M, major allele; m, minor allele.
* Estimates are adjusted slopes (β) and 95 % in the given units per % n-3 LCPUFA in whole blood; insulin and TAG were ln-transformed, and their estimates are given on a logarithmic scale. n-3 LCPUFA is given as the sum of EPA and DHA (%) in whole blood.
† P for interaction between n-3 LCPUFA and PPARG2 rs180182 in linear mixed models adjusted for age, sex × puberty, height, BMI z-score, school and class.
‡ n 534 for MM and n 173 for Mm/mm.
Effect modification by APOE genotypes
As expected, compared with children with the most common genotype E3/E3, those with E3/E2 + E2/E2 had lower LDL-cholesterol (1·92 (sem 0·05) v. 2·35 (sem 0·03) mmol/l) and higher HDL-cholesterol (1·52 (sem 0·03) v. 1·44 (sem 0·01)), whereas children carrying E3/E4 + E4/E4 had higher LDL-cholesterol of 2·55 (sem 0·04) mmol/l (all P < 0·05). APOE genotype modified the association between FADS rs1535 and HDL-cholesterol (P rs1535 × APOE = 0·019), as rs1535 major allele homozygotes carrying APOE2 had higher HDL-cholesterol than all other genotype combinations (Fig. 3). There were no interactions between whole-blood n-3 LCPUFA and APOE genotype (Table 3). However, when looking at the two n-3 LCPUFA separately, DHA was positively associated with HDL-cholesterol only among children with APOE2 (P n-3 LCPUFA × APOE = 0·04, data not shown). The unadjusted models showed the same overall results (data not shown).
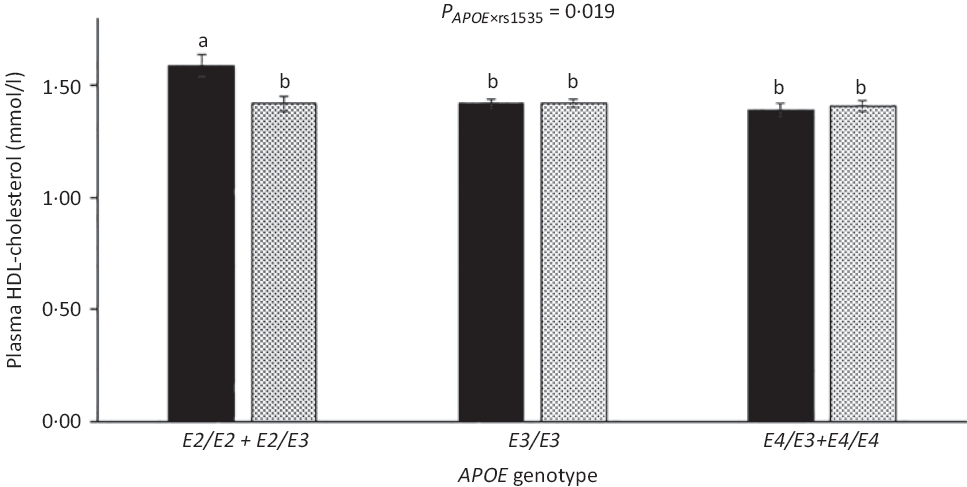
Fig. 3. HDL-cholesterol in the children according to fatty acid desaturase (FADS) rs1535 genotype and APOE genotype. Bars and error lines indicate raw means with their standard errors, respectively. P values are shown for rs1535 × APOE interaction. a,b Unlike letters indicate pairwise differences based on a linear mixed model adjusted for age, sex × puberty, height, BMI z-score, school and class (n 713). ![]() , rs1535 MM;
, rs1535 MM; ![]() , rs1535 Mm/mm.
, rs1535 Mm/mm.
Table 3. Associations between n-3 long-chain PUFA (LCPUFA) and the cardiometabolic markers according to APOE genotype*
(β Coefficients and 95 % confidence intervals)
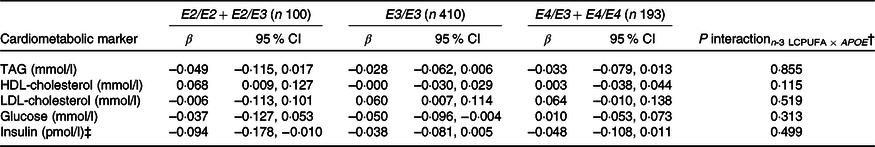
* Estimates are adjusted slopes (β) and 95 % per % n-3 LCPUFA in whole blood; insulin and TAG were ln-transformed, and their estimates are given on a logarithmic scale. n-3 LCPUFA is given as the sum of EPA and DHA (%) in whole blood. Twenty-one children with E2/E4 were excluded from the analysis.
† P for interaction between n-3 LCPUFA and APOE in linear mixed models adjusted for age, sex × puberty, height, BMI z-score, school and class.
‡ n 95 for E2/E2 + E2/E3, n 391 for E3/E3 and n 183 for E4/E3 + E4/E4.
Discussion
The present study showed that children carrying two FADS rs174448 major alleles, who have previously been shown to have higher blood n-3 LCPUFA(Reference Lauritzen, Sorensen and Harslof5), had lower plasma TAG and higher HDL-cholesterol than minor allele carriers. Major allele homozygotes of FADS rs1535 also had higher HDL-cholesterol, but only when also carrying APOE2. In contrast, associations between whole-blood n-3 LCPUFA and the cardiometabolic markers did not seem to depend on APOE genotype. There were no interactions between n-3 LCPUFA status and PPARA genotype, but n-3 LCPUFA was associated with lower TAG and insulin in carriers of PPARG2 minor allele, which has been shown to reduce transcription, and positively associated with LDL-cholesterol in major allele homozygotes. These data could indicate that endogenous as well as exogenous n-3 LCPUFA can modulate children’s cardiometabolic profile and that PPARγ2 and apoE play some role in their mechanisms of action, thereby suggesting that potential benefits of n-3 LCPUFA consumption may differ between children dependent on their genotype.
FADS rs174448 was associated with plasma TAG and HDL-cholesterol, and this was also indicated, but less pronounced, for rs1535. FADS catalyse the endogenous conversion of short- to long-chain PUFA of both the n-3 and n-6 family, and the SNP generally encode desaturase enzymes with reduced activity. In the children of the present cohort and among infants, we have seen that minor allele carriage of rs174488 was associated with low n-3 LCPUFA status and a tendency of low arachidonic acid (20 : 4n-6)(Reference Lauritzen, Sorensen and Harslof5,Reference Harslof, Larsen and Ritz6) , whereas minor allele carriage of rs1535 was only associated with low arachidonic acid in the children(Reference Lauritzen, Sorensen and Harslof5) and even with high DHA in the infants(Reference Harslof, Larsen and Ritz6). This indicates that the associations between rs174448, TAG and cholesterol in the present study could be driven by reduced n-3 LCPUFA status in the minor allele carriers. This would fit with results from numerous trials showing that fish oil supplementation lowers plasma TAG and increases HDL-cholesterol(Reference Abdelhamid, Martin and Bridges40). The estimated 0·04 mmol/l lower TAG and higher HDL-cholesterol in the rs174448 major allele homozygotes compared with minor allele carriers correspond to 50–100 % of the changes that we found in a recent randomised trial in 8–9-year-old children in response to intervention with oily fish several times per week(Reference Vuholm, Rantanen and Teisen41).
Few studies have investigated associations between FADS SNP, TAG and cholesterol in children. Standl et al.(Reference Standl, Lattka and Stach11) found that minor allele carriage of some other FADS SNP was associated with high plasma TAG and low HDL in 10-year-old German children. In contrast, in our infant study, rs174448 minor allele was not associated with TAG, but with lower LDL-cholesterol in all infants as well as with lower HDL-cholesterol among breastfed infants only(Reference Lauritzen, Amundsen and Damsgaard12). In line with our results, previous studies in adults have also reported associations between FADS minor alleles and higher TAG, as well as lower HDL-cholesterol(Reference Chasman, Pare and Mora42,Reference De Silva, Freathy and Palmer43) . We found that HDL-cholesterol was higher in children carrying APOE2 and two rs1535 major alleles compared with all other genotype combinations. n-3 LCPUFA may increase reverse cholesterol transport via activation of hepatic transcription factors(Reference Davidson44), and APOE2 has been associated with slower catabolism of HDL particles(Reference Blanchard, Ramin-Mangata and Billon-Crossouard45), which may result in higher circulating HDL-cholesterol when combined with higher LCPUFA. However, we did not find interaction between APOE genotype and n-3 LCPUFA on HDL-cholesterol or any other associations with the cardiometabolic markers, although DHA was positively associated with HDL-cholesterol only among children with APOE2, when the two n-3 LCPUFA were analysed separately.
Among the blood lipid outcomes, the PPARG2 SNP rs1801282 modified associations between n-3 LCPUFA and TAG and LDL-cholesterol. The observed positive association between n-3 LCPUFA and LDL-cholesterol among PPARG2 rs1801282 major allele homozygotes was not seen in our infant fish oil trial(Reference Harslof, Damsgaard and Hellgren2) or, to our knowledge, in previous trials in adults. For TAG, inverse associations were only seen in the minor allele carriers, which is in agreement with the PPARG2-dependent effects of fish oil supplementation in our previous infant study(Reference Harslof, Damsgaard and Hellgren2) and in studies in adults(Reference Lindi, Schwab and Louheranta46,Reference Binia, Vargas-Martinez and Ancira-Moreno47) . The rs1801282 minor allele has been shown to reduce transcription(Reference Masugi, Tamori and Mori48), and we hypothesise that increased levels of PPAR-γ2 ligands such as EPA and DHA may have a larger impact on the transcription of target genes (such as the lipoprotein lipase gene), when PPAR-γ2 is less efficient. Besides lipid metabolism, PPAR-γ is also important in the regulation of genes involved in glucose metabolism(Reference Ahmadian, Suh and Hah49) and animal studies have shown that n-3 LCPUFA may mediate effects on circulating glucose through PPAR-γ (Reference Wakutsu, Tsunoda and Shiba50,Reference Yu, Wu and Cheng51) . We found no interaction between FADS or n-3 LCPUFA on plasma glucose, but that rs1801282 modified the association between n-3 LCPUFA and serum insulin with an inverse association in minor allele carriers only. This concurs with the results of our previous infant trial, where fish oil reduced plasma glucose in carriers of rs1801282 minor allele only(Reference Harslof, Damsgaard and Hellgren2) and comparable interactions were indicated in a cross-sectional study among Finnish adults(Reference Ylonen, Salminen and Lyssenko52). Such a genotype-dependent effect of n-3 LCPUFA may explain why only some studies have shown effects of fish oil supplementation on glucose homoeostasis, but needs replication.
The magnitude of the estimated genotype-specific effect in the present study corresponds to a reduction in TAG and insulin of about 0·11 mmol/l and 8 pmol/l, respectively, among PPARG2 minor allele carriers and no change in major allele homozygotes, if all children’s whole-blood n-3 LCPUFA is increased from 3·5 to 5·0 %. This could be achieved by consumption of oily fish several times per week as demonstrated in our recent fish trial(Reference Vuholm, Teisen and Buch53). If true, such a genotypic difference would likely be of relevance for long-term cardiometabolic health. Our results did not indicate that the PPARA SNP rs1800206 was a potential effect modifier. Previous cross-sectional studies in adults have shown that rs1800206 modified the association between total PUFA intake and plasma TAG(Reference Tai, Corella and Demissie26) and HDL-cholesterol(Reference Chan, Tan and Deurenberg-Yap28). We cannot rule out the possibility that the low minor allele frequencies of rs1800206 and limited sample size of the present study may have compromised our ability to detect any effect modifications by this SNP.
The present study is limited by its cross-sectional design, which does not allow for inferences about causality, and by the reduced power in especially the genotype interaction analyses. The study is merely hypothesis generating and needs replication in larger studies. It is however strengthened by the targeted approach, the considerable sample size and the use of the whole-blood n-3 LCPUFA biomarker, which reflects dietary intake of recent weeks(Reference Metherel, Armstrong and Patterson54). Due to the exploratory nature of the study and since the outcome variables correlated, we did not adjust for multiple testing, but interpreted our findings with caution and focus on consistency, and the results were highly consistent in adjusted and unadjusted analyses. The original power calculation for the study was based on a metabolic syndrome score assessed after a school meal intervention, so some of the results could be chance-findings. Due to the low number of minor allele homozygotes, we pooled the minor allele homozygotes and heterozygotes in the analyses, which is a commonly used approach(Reference Standl, Lattka and Stach11), even without certainty that the minor allele was acting in a dominant fashion, that is, that the number of minor alleles is less relevant. Although genotypes are less prone to confounding than, for example, measures of dietary intake, they may be confounded by genetic ancestry, which we did not correct for. However, this is unlikely to be a major confounder in our study, since the study population was highly homogeneous with 95 % of the children being of white, Caucasian origin. Another potential confounder, which we did not control for, is socio-economic status; however, as previously described(Reference Damsgaard, Eidner and Stark29), most of the children came from households with a higher education. Finally, the study sample was to a large degree representative of Danish children of this age(Reference Damsgaard, Eidner and Stark29) and there were no apparent differences between the children, who were included and not included, in the present study. However, other studies would be needed to verify if the demonstrated associations could be found among populations with different demographics such as ethnic backgrounds, rates of overweight and dietary habits.
In conclusion, this study among Danish children showed that the least common allele of FADS rs174448 was associated with high TAG and low HDL-cholesterol, whereas those with two FADS rs1535 major alleles had high HDL-cholesterol, but only if they were APOE2 carriers. APOE genotype did not modify the beneficial associations between n-3 LCPUFA and TAG, LDL-cholesterol and insulin, but these associations were mainly seen in children with the variant genotype of PPARG2. These results indicate that (n-3) LCPUFA may affect children’s cardiometabolic profile in a potential genotype-specific manner, which may suggest that some children could benefit more from n-3 LCPUFA consumption than others. However, due to the cross-sectional design and limited power, the results need replication in larger genetic studies and in randomised trials, which examine the combined effect of n-3 LCPUFA and genotype on children’s cardiometabolic health.
Acknowledgements
We gratefully thank the participating children and their families.
The work was supported by Nordea-fonden, Denmark (grant no. 02-2010-0389).
C. T. D., S. V. and L. L. designed the research; C. T. D. led the data collection; K. D. S. was responsible for the fatty acid analyses; S. V. performed the statistical analysis; C. T. D. wrote the paper and had primary responsibility for the final content. All authors read and approved the final manuscript.
None of the authors has any conflicts of interest to declare.
Supplementary material
For supplementary material referred to in this article, please visit https://doi.org/10.1017/S0007114520002822









