Analysis of the human intestinal microbiota has rapidly evolved during the past 10 years (Tannock et al. Reference Tannock, Munro, Harmsen, Welling, Smart and Gopal2000). Molecular techniques, based on detecting nucleotide sequences of 16S rRNA using fluorescent in situ hybridisation (FISH) and PCR amplification combined with separation by denaturing gel electrophoresis, have enabled a good assessment of the composition of the microbiota and the complexity of the dominant species present (Blaut et al. Reference Blaut, Collins, Welling, Dore, Van Loo and De Vos2002). Moreover, these methods provide an opportunity to estimate the transient presence of food-borne bacteria within the predominant intestinal bacterial population (Alander et al. Reference Alander, Satokari, Korpela, Saxelin, Vilpponen-Samena, Mattila-Sandholm and von Wright1999; Tannock et al. Reference Tannock, Munro, Harmsen, Welling, Smart and Gopal2000; Rochet et al. Reference Rochet, Rigottier-Gois and Sutren2006).
Intestinal microbiota can induce both positive and negative effects on host physiology (Gorbach, Reference Gorbach1986; Macfarlane & Cummings, Reference Macfarlane, Cummings, Phillips, Pemberton and Shorter1991) and there has been great interest in the feasibility of modulating intestinal microbiota in a beneficial way in order to improve health. In this field, lactic acid bacteria with probiotic properties have been considered as potentially useful (Shortt, Reference Shortt1999; Teitelbaum & Walker, Reference Teitelbaum and Walker2002; Guarner et al. Reference Guarner, Perdigon, Corthier, Salminen, Koletzko and Morelli2005). Lactic acid bacteria have been used traditionally in food fermentation for thousands of years. Yogurt is milk fermented using a combination of Lactobacillus delbrueckii ssp. bulgaricus and Streptococcus salivarius ssp. thermophilus. More recently, other bacteria, mainly lactobacilli or bifidobacteria, have been combined with yogurt starters to produce fermented milks with specific functional properties related to health. Fermented milks deliver a large number of lactic acid bacteria into the gastrointestinal tract. These micro-organisms are capable of partially resisting gastric and bile secretions in vitro and in vivo (Mater et al. Reference Mater, Bretigny, Firmesse, Flores, Mogenet, Bresson and Corthier2005) and can deliver enzymes and other substances into the intestines (Marteau & Rambaud, Reference Marteau and Rambaud1993; Oozeer et al. Reference Oozeer, Goupil-Feuillerat, Alpert, van de Guchte, Anba, Mengaud and Corthier2002, Reference Oozeer, Furet, Goupil-Feuillerat, Anba, Mengaud and Corthier2005). The ability of yogurt to provide β-galactosidase and reduce symptoms of lactose intolerance has been well documented (Marteau & Rambaud, Reference Marteau and Rambaud1993; Sanders, Reference Sanders1993). Lactic acid bacteria have also been claimed to modify the intestinal environment. Some studies have shown that L. acidophilus, L. casei and Bifidobacterium bifidum can decrease potentially harmful enzymes of the resident microbiota such as β-glucuronidase and nitroreductase (Goldin, Reference Goldin1986; Marteau et al. Reference Marteau, Flourie, Pochart, Chastang, Desjeux and Rambaud1990; Guerin-Danan et al. Reference Guerin-Danan, Chabanet, Pedone, Popot, Vaissade, Bouley, Szylit and Andrieux1998). These results were obtained during clinical trials involving volunteers who for a few weeks consumed a controlled diet containing fermented milk, this period of consumption being compared with an initial period without these milk products.
The objective of the present study was to compare the composition and metabolism of the faecal microbiota in human subjects who regularly consumed at least 200 g yogurt per d or no yogurt. The effects of consuming dairy products on the dominant intestinal microbiota were evaluated using molecular approaches: in situ hybridisation and PCR amplification combined with separation by denaturing gel electrophoresis. Micro-organisms from fermented milk were monitored in faecal samples using specific primers. The following metabolic characteristics of the faecal microbiota were studied: glycosidases involved in carbohydrate hydrolysis (α- and β-galactosidase and β-glucosidase) and in mucin degradation (neuraminidase, N-acetyl-galactosaminidase and α-l-fucosidase), β-glucuronidase, nitrate and azoreductase activities as well as bile acid and neutral sterol microbial conversions. SCFA, NH3 and iso-acids were also investigated as markers of glycolytic and proteolytic fermentation.
Materials and methods
Subjects
Fifty-one healthy subjects living in the Paris area were included in the study. They were selected from the participants in the SU.VI.MAX cohort (Hercberg et al. Reference Hercberg, Preziosi, Briancon, Galan, Triol, Malvy, Roussel and Favier1998, Reference Hercberg, Galan, Preziosi, Bertrais, Mennen, Malvy, Roussel, Favier and Briancon2004), who were providing regular information on their dietary intake by completing at least ten 24-h record questionnaires over a period of 2·5 years. All subjects consumed their usual diet, with group Y containing subjects declaring an intake of 200–400 g yogurt per d (246 (se 7)) and group N, non-consumers. Group Y included thirty individuals (aged 35–60 years, mean age 46 years, twelve males and eighteen females) while group N included twenty-one individuals (aged 35–60 years, mean age 47 years, nine males and twelve females), because of dietary habits in the Paris area where most people are used to consuming yogurt. In group Y, the subjects regularly consumed soft white cheese (fromage blanc, 97 (se 22) g/d) and in group N, only occasionally (23 (se 26) g/d) (P = 0·032). No differences were observed concerning the consumption of milk (82 (se 21) and 67 (se 25) g/d in groups Y and N, respectively, P = 0·642) or cheese (46 (se 10) and 38 (se 11) g/d in groups Y and N, respectively, P = 0·594).
Stools were collected once by each subject in sterile plastic containers and immediately frozen at − 20°C for subsequent analyses.
PCR-temporal temperature gradient gel electrophoresis
Total DNA was extracted as described previously (Suau et al. Reference Suau, Bonnet, Sutren, Godon, Gibson, Collins and Dore1999) from 0·25-g aliquots of homogenised faeces or 2·0-g aliquots of yogurt and four fermented milks widely consumed in France. The concentration and integrity of nucleic acids were determined visually by electrophoresis on agarose (10 g/l) gel containing ethidium bromide. Forward U968-GC (5′-CGC CCG GGG CGC GCC CCG GGC GGG GCG GGG GCA CGG GGG GAA CGC GAA GAA CCT TAC-3′) and reverse T1401 (5′-GCG TGT GTA CAA GAC CC-3′) primers were used to amplify the V6 to V8 regions of bacterial 16S rDNA (Zoetendal et al. Reference Zoetendal, Akkermans and De Vos1998). Primers Lac1 (5′-AGC AGT AGG GAA TCT TCC A-3′) and Lac2GC (5′-CGC CCG CCG CGC GCG CGG CGG GCG GGG GCG GGG GCG GGG CAC GGG GGG ATT YCA CCG CTA CAC ATG-3′) were used for specific amplification of the 16S rDNA sequences of Lactobacillus, Pediococcus, Leuconostoc and Weissella genera, belonging to the Lactobacillus group (Walter et al. Reference Walter, Hertel, Tannock, Lis, Munro and Hammes2001). These two sets of primers produced 433 and 327 bp PCR amplicons, respectively. All primers were purchased from Qbiogene Research Services (Evry, France). For the first set of primers, the PCR conditions were as described previously (Seksik et al. Reference Seksik, Rigottier-Gois, Gramet, Sutren, Pochart, Marteau, Jian and Dore2003). For the Lac1 and Lac2GC primers, the hybridisation temperature was 61°C for 1 min. PCR products were analysed by electrophoresis on agarose (10 g/l) gel containing ethidium bromide to check their size (500 bp) and estimate their concentration. The DCode Universal Mutation Detection System (Bio-Rad, Paris, France) was used for the sequence-specific separation of PCR products, as previously described (Seksik et al. Reference Seksik, Rigottier-Gois, Gramet, Sutren, Pochart, Marteau, Jian and Dore2003). At the beginning of electrophoresis, for better resolution, the voltage was fixed at 20 V for 15 min. Then, for the first set of amplicons, electrophoresis was run for 16 h at 64 V with initial and final temperatures of 66 and 70°C, respectively (ramp rate 0·2°C per h). For amplicons obtained with primers Lac1 and Lac2GC, the initial and final temperatures were 63.8 and 70°C, respectively (ramp rate 0·4°C per h), at a voltage of 66 V. Each well was loaded with 100–200 ng amplified DNA plus an equal volume of gel loading dye (bromophenol blue (0·5 g/l)–xylene cyanol (0·5 g/l)–glycerol (700 g/l)). For each gel, three lanes were loaded with a marker obtained by mixing the PCR products of seven rDNA clones obtained from an adult male faecal sample (Suau et al. Reference Suau, Bonnet, Sutren, Godon, Gibson, Collins and Dore1999). Clones were related to members of the Clostridium coccoides group, Clostridium leptum subgroup, Bacteroides group and Bifidobacterium genus. After the completion of electrophoresis, the gels were stained in the dark by immersion for 30 min in a solution of SYBR Green I Nucleic Acid Gel Stain (Roche Diagnostics, Meylan, France) and read on a Storm system (Molecular Dynamics, Amersham Biosciences, Orsay, France).
Fluorescent in situ hybridisation combined with flow cytometry
Whole cell FISH was performed as previously described (Rigottier-Gois et al. Reference Rigottier-Gois, Le Bourhis, Gramet, Rochet and Dore2003) except that fixation was performed using frozen–thawed rather than fresh samples, within 1 month of collection (Rochet et al. Reference Rochet, Rigottier-Gois, Rabot and Dore2004). The group-specific probes used are presented in Table 1. Data acquisition was performed using a Facs Calibur flow cytometer (Becton Dickinson, San Jose, CA, USA) equipped with an air-cooled Ar ion laser providing 15 mW at 488 nm combined with a 635 nm red diode laser (Rigottier-Gois et al. Reference Rigottier-Gois, Le Bourhis, Gramet, Rochet and Dore2003). Subsequent analyses were conducted using CellQuest software (Becton Dickinson). Results were expressed as percentages of cells hybridising with the different group-specific probes relative to total bacteria hybridising with the EUB 338 probe, after the elimination of background fluorescence as determined using the NON 338 probe. Based on previous experience where faecal samples were added with serial dilutions of pure strains, the probes enabled the reliable detection of added target cells down to 0·5 % of the total. However, when values below 0·5 % were returned by the software, they were kept unchanged for the purposes of calculation.
Table 1 Probes used for the fluorescent in situ hybridisation analysis of intestinal microbiota*

* For details of procedures, see p. 127.
OPD, oligonucleotide probe database.
Biochemical analyses
Enzyme activities were measured as previously described (Andrieux et al. Reference Andrieux, Membre, Cayuela and Antoine2002) using a thermo-regulated anaerobic chamber (H2–CO2–N2 (10:10:80, by vol.)). Frozen–thawed faecal samples were diluted 1:20 using pre-reduced PBS (pH 6·7). α- and β-galactosidase, β-glucosidase, β-glucuronidase, N-acetyl-galactosaminidase and α-l-fucosidase activities were measured by determining the rate of p-nitrophenol release from p-nitrophenyl glycosides. Azoreductase activity was determined using amaranth as the substrate. Neuraminidase activity was measured using 4-methylumbelliferyl-N-acetylneuraminic acid as the substrate. Nitrate reductase was determined by the generation of nitrite. Enzyme activities were expressed as μmol metabolised substrate per min and per g protein.
Protein concentrations were determined in triplicate using the method described by Lowry et al. (Reference Lowry, Rosebrough, Farr and Randall1951) with 1:500 faecal dilutions in Na2CO3 (20 g/l) and NaOH (0·1 m). Bovine serum albumin was used as the standard.
SCFA concentrations in faecal samples were analysed in duplicate after water extraction of acidified samples using GC (Perkin-Elmer 1020 GC; Perkin-Elmer, Saint Quentin, France) (Andrieux et al. Reference Andrieux, Membre, Cayuela and Antoine2002). NH3 was determined using the Berthelot method adapted by Drospy & Boy (Reference Drospy and Boy1961).
For bile acid analysis, the continuous extraction of 4-g aliquots of homogenised faeces was carried out for 48 h using boiling ethanol (100 ml) in a Soxhlet apparatus (Riottot et al. Reference Riottot, Olivier, Huet, Caboche, Parquet, Khallou and Lutton1993). A 4-ml aliquot of the ethanol extract was hydrolysed by adding 1 ml 10 m-NaOH and heating the alkaline mixture to 120°C for 3 h under pressure. The hydrolysate was then cooled to room temperature, diluted with an equal volume of water (5 ml) and neutral sterols were extracted with 3 × 10 ml petroleum ether. The delipidated aqueous phase was then placed on ice, acidified to pH 2·0 with concentrated HCl and bile acids were extracted with 3 × 10 ml ethylic ether. The combined solvent fractions containing either neutral or acidic sterols were evaporated to dryness and the samples were reconstituted in 2 ml hexane or methanol, respectively, and analysed by GLC, taking 5α-cholestane as the external standard.
Statistical analyses
PCR-temporal temperature gradient gel electrophoresis (TTGE) profiles were compared using GelCompar software (GelComparII™ version 3.5; Applied Maths, Sint-Martens-Latem, Belgium). Analysis included calculation of the number, position and intensity of PCR-TTGE bands (PCR-amplified 16S rDNA fragments) in the gel. Pearson correlation provided similarity based upon densitometric curves. Pearson similarity coefficients were calculated to generate a similarity matrix. The dendrogram was obtained from this matrix using the Unweighted Pair Group Method using Arithmetic averages algorithm. Cell proportions as estimated by FISH and biochemical data were analysed using the general linear model procedure and the MEAN and LSMEAN statements in SAS Version 8.2 (SAS Institute, Cary, NC, USA) with one two-level factor for comparison between yogurt consumers and non-consumers. Results were expressed as means and their standard errors and were considered significant at P ≤ 0·05. All data were examined for normal distributions and log-transformed when necessary prior to analysis. In this case, means were back-transformed for reporting as estimated means, and standard errors were estimated by assuming that standard errors on the logarithmic scale approximately equal CV on the actual scale. We employed Fisher's R-to-z transformation and the CORR procedure under SAS to test whether correlations between yogurt consumption and different variables were significantly different from zero.
Results
Composition of faecal microbiota
Fluorescent in situ hybridisation analysis
Using FISH adapted for detection by flow cytometry, eight phylogenetic groups were investigated in faecal samples from yogurt consumers (Y) and non-consumers (N) (Fig. 1). The only significant difference concerned Enterobacteriaceae, which were found at significantly lower levels in group Y (0·3 (SE 0·3) % v. 1·6 (SE 0·4) % in group N, P = 0·006). They were below the threshold of detection (0·5 %) in twenty-six out of thirty yogurt consumers, and between 0·5 and 2·4 % in the remaining four. Among non-consumers, the levels of Enterobacteriaceae were below the detection threshold in twelve subjects while it ranged from 4·8 to 7·2 % in four subjects. Interestingly, within group Y, the proportion of bifidobacteria was positively correlated with fermented milk consumption (r 0·43, P = 0·018). Overall, the cumulative proportions of cells hybridised with the eight group-specific probes among bacteria detected using the EUB 338 probe averaged 68·2 (SE 1·9) %, without any significant difference between groups Y and N.
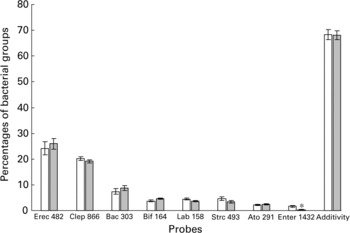
Fig. 1 Percentages of bacterial groups obtained by FISH analysis (see details in Table 1) in faecal samples from non-consumers (Group N, □) and consumers of yogurt (Group Y, ![]() ). Values are estimated means with their standard errors. *Indicates significant difference (P = 0·006) between mean values.
). Values are estimated means with their standard errors. *Indicates significant difference (P = 0·006) between mean values.
PCR-temporal temperature gradient gel electrophoresis analysis
PCR-TTGE profiles obtained with general primers revealed the same broad inter-individual variability (Pearson similarity coefficient < 80 %) in both groups of subjects, without any significant differences between yogurt consumers and non-consumers. No band migrated at the same level as bands obtained from fermented milk (results not shown).
PCR-TTGE profiles generated with Lac1 and Lac2GC primers displayed no bands in three samples from group N, while one to six bands were observed in the forty-eight remaining samples from groups N and Y (Fig. 2). One band co-migrating with that obtained from L. bulgaricus DNA was observed in 73 % and 28 % of samples from groups Y and N, respectively (P = 0·003). The PCR-TTGE profiles obtained with yogurt, three fermented milks and ‘fromage blanc’ revealed one band migrating at the same level (Fig. 3). Identity of L. bulgaricus was confirmed by sequencing the excised band from the gels of six yogurt consumers and from fermented milks. One band co-migrating with that obtained from L. casei DNA was obtained in four samples from group Y and one sample from group N; L. bulgaricus was found together with L. casei in these profiles and in the profile of the fermented milk Actimel® (Danone, France). One band was identified as Leuconostoc mesenteroides in the fermented milk Yorik® (Yoplait, France) and in three samples from groups Y and N. It is also interesting to note that TTGE analysis of fermented milks revealed a weaker band for the probiotic organism than for the starter organism.
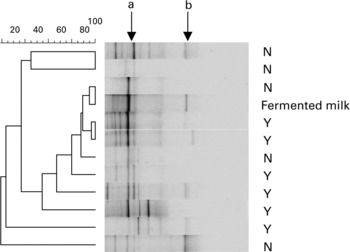
Fig. 2 PCR-TTGE profiles obtained with Lac1 and Lac2GC primers from faecal samples of non-consumers (N) and consumers (Y) of yogurt and from fermented milk containing yogurt starters and L. casei (Actimel®, Danone, France). Arrows show L. delbrueckii subsp. bulgaricus (a) and L. casei (b).

Fig. 3 PCR-TTGE profiles obtained with Lac 1 and Lac2GC primers from six dairy products, and from L. delbrueckii subsp. bulgaricus (a) and L. casei (b). M: marker obtained by mixing PCR products of seven rDNA clones (Suau et al. Reference Suau, Bonnet, Sutren, Godon, Gibson, Collins and Dore1999).
Metabolic activities
Among the nine metabolic bacterial enzyme activities investigated, the only significant difference concerned β-galactosidase, which was found at significantly higher levels in group Y (Table 2). It was higher than 100 μmol/min per g protein in five yogurt consumers and higher than 200 μmol/min per g protein in two of them. Among non-consumers, this activity only reached 100 μmol/min per g protein in one subject while it was inferior to 50 μmol/min per g protein in five subjects. Moreover, in group Y, β-galactosidase activity was positively correlated with the daily consumption of yogurt (r 0·66, P < 0·0001).
Table 2 Bacterial enzyme activities (μmol/min per g protein) in faecal samples from non consumers (group N) or consumers (group Y) of yogurt* (Mean values with their standard errors)

* For details of subjects and procedures, see pp. 127-128.
The overall concentration of SCFA was about 100 μmol/g fresh faeces in both groups of subjects (P = 0·151) without any significant inter-group difference in composition (Table 3). However, in group Y, 67 % of values were superior to 100 μmol/g compared with 38 % in group N. Moreover, the lowest values ( < 40 μmol/g) were all found in group N.
Table 3 Concentration of SCFA (μmol/g fresh stools) in faecal samples from non-consumers (group N) or consumers (group Y) of yogurt* (Mean values with their standard errors)

* For details of subjects and procedures, see pp. 127-128.
Cholic and chenodeoxycholic acids are the two primary bile acids (i.e. synthesised by the liver) in man and physiologically are poorly represented in stools. Their respective proportions were less than 5 % (mass percentage of total faecal bile acids, non-significant difference) in both groups of subjects (Table 4). All other bile acids are secondary bile acids, i.e. bacterial metabolites of cholic and chenodeoxycholic acids. They did not differ significantly between groups, except for 7-keto-lithocholic acid, which was found at lower levels (P = 0·012) in yogurt consumers (Table 4). Concomitantly, the proportion of ursodeoxycholic acid, which is the 7β-OH epimer of chenodeoxycholic acid with 7-keto-lithocholic acid as the intermediate reaction product, tended to be lower (P = 0·078) in yogurt consumers. But the overall proportion of secondary bile acids did not significantly differ between groups (Table 4). Untransformed neutral sterols of animal and plant origin, i.e. cholesterol and β-sitosterol, each represented about 10 % total neutral sterols in stools, without there being any significant difference between groups (Table 4). Nor were bacterial products of cholesterol (coprostanol, cholestanol and coprostanone) and β-sitosterol (copro-β-sitosterol) modified by yogurt consumption (Table 4).
Table 4 Profiles of acidic and neutral sterols (mass percentages of total bile acids and neutral sterols) in faecal samples from non-consumers (Group N) and consumers (Group Y) of yogurt¶ (Mean values with their standard errors)

* Bacterial metabolites of cholic acid.
† Bacterial metabolites of chenodeoxycholic acid.
‡ Deoxycholic + ursocholic +7-keto-deoxycholic + litocholic + ursodeoxycholic +7-keto-lithocholic.
§ Bacterial metabolites of cholesterol.
‖ Bacterial metabolite of β-sitosterol.
¶ For details of subjects and procedures, see pp. 127-128.
Discussion
During the present study, molecular and biochemical approaches were used to assess the effects of fermented milk consumption when subjects were consuming self-chosen diets. The human volunteers were selected from participants in the SU.VI.MAX cohort (Hercberg et al. Reference Hercberg, Galan, Preziosi, Bertrais, Mennen, Malvy, Roussel, Favier and Briancon2004). They either consumed at least 200 g yogurt per d or no fermented milk products at all. Their food records did not include the brand of the foods consumed; we therefore compared the profiles of faecal bacteria from subjects with those of the commercial products generally consumed in the Paris area where the subjects lived.
Using group-specific primers for Lactobacillus and related genera with sequencing confirmation, L. delbrueckii ssp. bulgaricus was identified in four out of five commercial fermented milks, including yogurt, and in ‘fromage blanc’, which was also widely consumed in group Y. Using the same group-specific primers (which normally detect Lactobacillus species present at a level of >106 colony-forming units/g wet faeces), L. delbrueckii ssp. bulgaricus was also identified in 73 % of faecal samples from yogurt consumers but in only 28 % of faecal samples from non-consumers. L. casei was also identified in three times more yogurt consumers than non-consumers. Conversely, using non-specific primers (which detect the 90–99 % most numerous species in the community) did not allow us to highlight any faecal bacteria from fermented milk in any subjects. These results suggest that, in most yogurt consumers, L. delbrueckii ssp. bulgaricus was present at a level where biological effects could be expected (Marteau & Rambaud, Reference Marteau and Rambaud1993). This was also the case in a reduced proportion of non-consumers, all but one of whom were the highest consumers of cheese (but not of ‘fromage blanc’) within their group. Consumption of L. bulgaricus-containing cheeses, such as Swiss (Emmental, Gruyère) or Mediterranean (Feta, Mozzarella, Provolone, etc.), could therefore explain the occurrence of this micro-organism in group N. But it should be emphasised that the current results do not provide information on the survival of this micro-organism in faeces and a fortiori during transit in the gastrointestinal tract, since PCR-amplified DNA detection does not distinguish between live, injured and dead cells. Other studies have provided evidence that L. delbrueckii ssp. bulgaricus and L. casei, known to exhibit poor survival when challenged in vitro by gastric acidity and bile salts, partly survive transit through the gastrointestinal tract of human volunteers (Pochart et al. Reference Pochart, Dewit, Desjeux and Bourlioux1989; Bouhnik, Reference Bouhnik1993; Yuki et al. Reference Yuki, Watanabe, Mike, Tagami, Tanaka, Ohwaki and Morotomi1999; Callegari et al. Reference Callegari, Morelli, Ferrari, Cobo Sanz and Antoine2004; Mater et al. Reference Mater, Bretigny, Firmesse, Flores, Mogenet, Bresson and Corthier2005) and Göttingen minipigs (Lick et al. Reference Lick, Drescher and Heller2001) ingesting yogurt. Furthermore, there is recent evidence that L. casei initiates protein synthesis during its transit in the digestive tract of mice harbouring human intestinal microbiota (Oozeer et al. Reference Oozeer, Goupil-Feuillerat, Alpert, van de Guchte, Anba, Mengaud and Corthier2002, Reference Oozeer, Furet, Goupil-Feuillerat, Anba, Mengaud and Corthier2005) and that some protective effects of probiotics, such as immuno-stimulation, are mediated by their own DNA rather than by their ability to survive through the gastrointestinal tract (Rachmilewitz et al. Reference Rachmilewitz, Katakura and Karmeli2004).
FISH analysis of the predominant bacterial groups in faeces showed that Enterobacteriaceae, which include pathogenic bacteria, were significantly less represented in yogurt consumers. Otherwise, chronic consumption of yogurt did not significantly alter other bacterial groups. But interestingly, within group Y, values obtained for the Bifidobacterium genus, which is recognised for its potential health-promoting capacities (Gibson et al. Reference Gibson, Beatty, Wang and Cummings1995), was positively correlated with fermented milk consumption. Taken altogether, these novel results suggest that a regular consumption of yogurt in a self-chosen diet could induce modifications in the indigenous microbiota.
During the present study, β-galactosidase activity in faeces was found to be significantly increased in yogurt consumers and to exhibit a positive correlation with the amount of yogurt consumed within this group. In yogurt, S. salivarius ssp. thermophilus contains a β-galactosidase, which is active in the digestive tract of germ-free mice (Drouault et al. Reference Drouault, Anba and Corthier2002), while L. delbrueckii ssp. bulgaricus does not (Pochart et al. Reference Pochart, Dewit, Desjeux and Bourlioux1989). This suggests that S. salivarius ssp. thermophilus may survive intestinal transit, even though overall levels of the streptococcus group did not significantly differ between subject groups. Other intestinal bacteria may also participate in lactose fermentation (He et al. Reference He, Priebe, Vonk and Welling2005), e.g. bifidobacteria, the proportion of which presented a positive correlation with the amount of fermented milk consumed in group Y. Otherwise, chronic consumption of yogurt did not significantly alter other enzyme activities. This is in contrast with the reports of authors who have found that some probiotics, mainly lactobacilli or bifidobacteria, may improve bacterial metabolism by decreasing potentially harmful enzymes such as β-glucuronidase, reductases and 7α-dehydroxylase (Rowland, Reference Rowland and Fuller1992; Roos & Katan, Reference Roos and Katan2000). Because the SU.VI.MAX questionnaire did not allow a distinction between subjects who consumed traditional yogurts (fermented with S. thermophilus and L. delbrueckii ssp. bulgaricus starter cultures) and those who consumed yogurts enriched with other probiotic bacteria, it cannot be excluded that the differences between groups were only small because the study participants mainly consumed traditional yogurts.
Sterol metabolism was weakly affected by yogurt consumption, as inferred from the respective proportions of the different molecular species of neutral and acidic sterols in the stools of yogurt consumers and non-consumers. It has been argued that the bile salt hydrolase activity of ingested bacteria, such as lactobacilli and bifidobacteria, might favour the early deconjugation of bile acids in the small intestine and their subsequent biotransformation by the resident microbiota, leading to the increased formation of cytotoxic secondary bile acids (Marteau et al. Reference Marteau, Gerhart, Myara, Bouvier, Trivin and Rambaud1995). This was not observed in the present study, where the overall percentages of secondary bile acids and of the main secondary lithocholic and deoxycholic acids, which are known to be the most cytotoxic, were not increased in yogurt consumers. The only difference concerned the minor secondary 7-keto-lithocholic acid, and to a lesser extent ursodeoxycholic acid, which were or tended to be decreased in yogurt consumers. This suggests that 7β-epimerisation of chenodeoxycholic to ursodeoxycholic acid, via 7-keto-lithocholic acid as the intermediate product (Lepercq et al. Reference Lepercq, Gérard, Béguet, Grill, Relano, Cayuela and Juste2004) could be decreased by long-term yogurt consumption. It is impossible to speculate as to the mechanisms involved or any impact on health.
In conclusion, the present work demonstrated the presence of food-borne bacteria in faeces from healthy volunteers who regularly included yogurt in their everyday diet in an industrialised country. It also suggests that the consumption of yogurt may ensure some advantageous changes to the equilibrium and metabolic activities of the indigenous microbiota.
Acknowledgements
We are grateful to C. Girard for her technical assistance, to O. David for statistical advice, to Dr. G. Corthier, Dr P. Langella and Dr. S. Rabot for helpful suggestions regarding the manuscript and to V. Hawken for her advice on English translation.









