Collagen is the most abundant protein in the body, comprising about one-third of total proteins. Collagen is contained in typical diets consisting of animal products such as meat, bone, tendon or fish skin( Reference Eastoe 1 ). Collagen extracted from collagen-rich materials with hot water is known as gelatin. Gelatin and its partial hydrolysate (collagen peptide (CP)) are widely used as food supplements. Ingestion of collagen has several beneficial effects, including increased bone mineral density( Reference Koyama, Hirota and Mori 2 , Reference Wu, Fujioka and Sugimoto 3 ), muscle strength( Reference Zdzieblik, Oesser and Baumstark 4 ), attenuation of joint pain and disability( Reference Walrand, Chiotelli and Noirt 5 , Reference Schauss, Stenehjem and Park 6 ), as well as increased diameter and density of collagen fibrils in the tendon and skin( Reference Minaguchi, Koyama and Meguri 7 , Reference Matsuda, Koyama and Hosaka 8 ).
The beneficial effects of collagen ingestion are not restricted to collagen-rich tissues such as the skin, bone or tendon. Collagen ingestion suppresses the activity of angiotensin I-converting enzyme, resulting in decreased blood pressure( Reference Saiga, Iwai and Hayakawa 9 , Reference Herregods, Van Camp and Morel 10 ). Ingestion of collagen also reportedly modulates blood lipid profiles in rats( Reference Wu, Fujioka and Sugimoto 3 , Reference Ratnayake, Sarwar and Laffey 11 , Reference Lin, Zhang and Yu 12 , Reference Koyama and Kusubata 14 ), mice( Reference Oliveira, Portugal and Cara 13 ) and rabbits( Reference Terpstra, Hermus and West 15 ); TAG and/or total cholesterol levels are markedly reduced in these animals when fed a CP-containing diet. It has been expected that the ingestion of collagen, which has been shown to elicit effects in various parts of the body( Reference Koyama, Hirota and Mori 2 – Reference Terpstra, Hermus and West 15 ), likely affects the liver, which is involved in various biological functions including lipid metabolism, detoxification, protein synthesis, hormone production and production of biochemicals necessary for digestion( Reference Tsung, Geller and Colnot 16 ). However, there are a few reports on the effects of collagen ingestion in the liver. In addition, even though the liver is involved in various biological functions, the condition of the liver is difficult to assess visually, unlike the skin or joints. Therefore, we focused on the effects of collagen ingestion on liver gene expression, and attempted to investigate this using a nutrigenomic approach using DNA microarray.
We have previously reported the effect of collagen ingestion on bone mineral density in mice( Reference Koyama, Hirota and Mori 2 ). In that study, the four diet conditions of 14 % casein, 10 % casein, 6 % casein+8 % gelatin, and 6 % casein+4 % gelatin were compared to investigate the effect of collagen ingestion under conditions of normal protein ratio diet (14 %) or moderately low-protein diet (10 %). Significant in vivo effects of gelatin ingestion were observed; mice fed a 6 % casein+4 % gelatin diet exhibited significantly higher bone mineral density than those fed a 10 % casein diet. In the present study, we used similar diets containing 10 % casein and 6 % casein+4 % CP (fish derived, MW 4000–6000, fragments of amino acid sequences of collagen that consist of – Gly-X-Y – triplets; the X and Y positions are frequently occupied by proline and hydroxyproline, respectively) to examine how collagen ingestion modulates gene expression in the liver, as determined by DNA microarray analysis. We also used the 14 % casein diet to confirm the biological state under the condition of normal protein ratio diet.
Methods
Animals and diets
Male 6-week-old BALB/cCrSlc mice were purchased from Japan SLC, Inc., and were acclimated to breeding conditions for 1 week in the laboratory animal facilities of Rakuno Gakuen University. The animals were divided into three groups (n 12 each) on the basis of body weight, so that the average body weight was equivalent among groups. Mice were housed individually under conventional conditions (20±2°C, 12 h light–12 h dark cycle) with free access to water and food. The basic powder diet AIN-93M containing 14 % protein (as casein) (14C) and the AIN-93M-based low-protein diets containing 10 % protein (as casein) (10C) or 6 % protein (as casein) plus 4 % protein (as CP, fish derived, MW 4000–6000; Nippi Inc.) (6C+4CP) were prepared by Oriental Yeast Co., Ltd (Table 1). Maize starch was used to compensate for the decreased weight of protein in the low-protein diets. Body weight as well as food and water intakes were measured once per week.
Table 1 Diet composition(%)
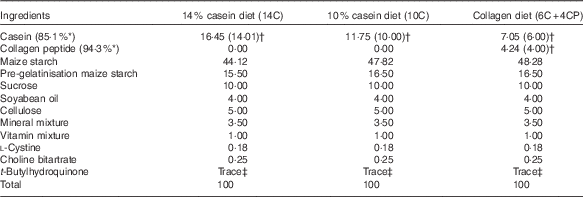
* Protein content of milk casein or collagen peptide.
† Values in parentheses indicate protein content of milk casein or collagen peptide in each diet.
‡ 0·0008 %.
After 10 weeks of rearing on the test diets, sampling was performed in the morning after a 3-h fasting period. After anaesthesia with 50 mg/kg sodium pentobarbital, blood was sampled from the right ventricle with a heparinised syringe, and plasma was stored at −80°C. For DNA microarray and Western blotting analyses, three mice exhibiting around median food efficiency (weight gain/food intake) were selected from each group. The whole liver was removed, and a portion of the first lobe (about 5×5×3 mm) was stored in RNAlater® (Ambion; Thermo Fisher) before isolation of total RNA. The residual liver was snap-frozen in N2 and stored at −80°C until used for biochemical analyses.
This study was conducted in accordance with the Act on Welfare and Management of Animals in Japan (Act No. 105 of 1 October 1973; partially revised on 22 June 2005), the Standards Relating to the Care and Management of Laboratory Animals and Relief of Pain (Notice No. 88 of the Ministry of the Environment, dated 28 April 2006), and the Guidelines for Methods of Animal Disposition Procedures (Prime Minister’s Office Notification No. 40, dated 4 July 1995; partially revised on 12 November 2007: Notice No. 105 of the Ministry of the Environment). The study protocol was approved by the Ethics Committee of Rakuno Gakuen University (No. VH23A8). We have read Animal Research: Reporting of In Vivo Experiments (ARRIVE) guidelines and confirmed that our experiments complied with these guidelines. The animal model and numbers in our study were decided after referring to other previous studies( Reference Koyama, Hirota and Mori 2 , Reference Oliveira, Portugal and Cara 13 ). In addition, the animal experiments were conducted in a blinded manner.
Biochemical analysis of blood
Biochemical analysis of blood plasma samples from all animals was entrusted to Nagahama Life Science Laboratory, Oriental Yeast Co., Ltd. The routine biochemical analyses were performed by absorbance measurements based on enzymatic methods using 7180 Clinical Analyzer (Hitachi High-Technologies Corporation). The panel of tests included the following: total cholesterol (T-CHO), free cholesterol (F-CHO), total protein, albumin (ALB), aspartate aminotransferase (AST; GOT), alanine aminotransferase (ALT; GPT), alkaline phosphatase, lactate dehydrogenase, leucine aminopeptidase, γ-glutamyl transferase, choline esterase, TAG, LDL-cholesterol, HDL-cholesterol, total bilirubin, total bile acid, pyruvic acid, total ketone bodies and glucose. The data on esterified cholesterol (E-CHO) were estimated by the calculation (T-CHO=F-CHO+E-CHO). In addition, the following items were measured by ELISA: adiponectin, insulin, leptin and insulin-like growth factor-1 (IGF-1).
Measurement of liver lipids
Determination of TAG as well as total and free cholesterol levels in liver samples from all animals was entrusted to Skylight Biotech, Inc. The lipid components of the liver samples were extracted using chloroform–methanol mixture solution, and determined by absorbance measurements based on enzymatic methods (reagent kits: Cholestest®TG or Cholestest®CHO; Sekisui Medical Co., Ltd and Cholescolor®Liquid FC; Toyobo Co., Ltd).
RNA preparation
Total RNA was isolated from each liver sample using ISOGEN® reagent (Nippon Gene Co.) and subsequently purified using an RNeasy® Mini Kit (Qiagen K.K.), according to the manufacturer’s protocol. Total RNA quality and quantity were assessed by agarose gel electrophoresis and spectrophotometry. Total RNA was used for the DNA microarray assay, as described below.
DNA microarray experiment
DNA microarray analysis was performed on liver samples from a total of six mice (three mice each from the 10C and 6C+4CP groups) by choosing average individuals from each group on the basis of their food efficiency values. For each sample, double-stranded complementary DNA (cDNA) was synthesised from 100 ng of purified total RNA, and biotinylated cRNA was transcribed from the cDNA using a GeneChip® 3’ IVT Express Kit (Affymetrix). The cRNA were subsequently fragmented and hybridised to a GeneChip® Mouse Genome 430 2.0 Array (Affymetrix) containing over 34 000 mouse genes. The arrays were washed and labelled with streptavidin–phycoerythrin using a GeneChip® Hybridization, Wash and Stain Kit and Fluidics Station 450 system (Affymetrix). Fluorescence was detected using a GeneChip® Scanner 3000 7G (Affymetrix). All experimental procedures were carried out according to the manufacturer’s instructions. All microarray data were Minimum Information About a Microarray Experiment (MIAME) compliant, and were deposited in a MIAME-compliant database – the National Center for Biotechnology Information Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/; GEO Series accession number GSE75171) – as detailed on The Functional Genomics Data Society website (http://fged.org/projects/miame/).
DNA microarray data analysis
Affymetrix GeneChip® Command Console software was used to reduce the array images to the intensity of each probe (CEL files). The CEL files were quantified using the Factor Analysis for Robust Microarray Summarization algorithm( Reference Hochreiter, Clevert and Obermayer 17 ) with the statistical packages R( 18 ) and Bioconductor( Reference Gentleman, Carey and Bates 19 ). Probe sets found to be differentially expressed between the 10C and the 6C+4CP groups were identified according to the Rank products method( Reference Breitling, Armengaud and Amtmann 20 ) using the statistical package R. The annotation file for the Mouse Genome 430 2.0 Array was downloaded from the Affymetrix website (30 March 2016, http://www.affymetrix.com/). Probe sets with a false-discovery rate (FDR) <0·05 were considered to reflect ingestion of CP. Gene-annotation enrichment analysis was then performed using the web tool Database for Annotation, Visualization, and Integrated Discovery (version 6.7)( Reference Huang da, Sherman and Lempicki 21 ) with Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) annotations. Expression Analysis Systematic Explorer (EASE) scores, which are modified Fisher’s exact test P-values( Reference Hosack, Dennis and Sherman 22 ), were used to evaluate statistically over-represented GO terms or KEGG pathways from the differentially expressed genes. Benjamini & Hochberg( Reference Benjamini and Hochberg 23 ) FDR corrections for multiple testing were used to correct the results. GO terms with FDR-corrected P-values of <0·05 were regarded as significantly enriched.
Real-time quantitative RT-PCR analysis
cDNA synthesis of total RNA (2 μg/sample) was carried out using High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher-Applied Biosystems) according to the manufacturer’s instructions. To perform real-time quantitative PCR, specific primers and probes for mouse PPARα, acyl-CoA oxidase 1, palmitoyl (Acox1), inositol-requiring enzyme 1 (IRE1) and ER degradation enhancer, mannosidase alpha-like 1 (Edem1) (as targets) and actin beta (Actb) as an endogenous control were purchased from Thermo Fisher-Applied Biosystems (TaqMan® Gene Expression Assays; ID: Mm00440939_m1, Mm01246834_m1, Mm00470233_m1, Mm00551797_m1, Mm02619580_g1, respectively). PCR was performed using 7300 Real-time PCR System (Thermo Fisher-Applied Biosystems) following the manufacturer’s protocol. The reaction solution for PCR was prepared in volumes of 20 μl/well containing 10 μl TaqMan® Universal PCR Master Mix (Thermo Fisher-Applied Biosystems), 1 μl primer and probe, and 9 μl cDNA in RNase-free water (500-fold dilution of the cDNA synthesis solution), and PCR was performed using two wells per sample. Amplification conditions were 2 min at 50°C, 10 min at 95°C and forty cycles each consisting of 15 s at 95°C and 1 min at 60°C. The standard curves were drawn by serial dilutions of the cDNA sample. The C t value obtained by amplification of each target was compared among the samples after normalisation using Actb expression levels.
Western blotting
Approximately 30 mg of each liver sample was excised on ice and homogenised in Tissue Protein Extraction Reagent (Thermo Fisher Scientific, Inc.) containing a 1/100 volume of Halt Protease and Phosphatase Inhibitor Cocktail (Thermo Fisher Scientific, Inc.). Each sample was then diluted with 2× sample buffer mix for SDS-PAGE (pH 6·8, 4 % SDS, 25 % glycerol, 125 mm-TRIS-HCl, 0·02 % bromophenol blue, 10 % 2-mercaptoethanol). After brief sonication and centrifugation (4°C, 1000 rpm, 10 min), the supernatant was collected and boiled at 100°C for 3 min. Each sample of 10 μlwas separated on an 8 % SDS-PAGE gel and transferred to a polyvinylidene difluoride membrane (Millipore). The membrane was blocked by incubation in TRIS-PBS (0·1 % Tween 20, PBS) containing 0·3 % skimmed milk for 30 min at room temperature. After blocking, the membrane was probed with rabbit antibodies against IRE1 or phospho-IRE1 (p-IRE1) (Abcam) and a rabbit antibody against glyceraldehyde-3-phosphate dehydrogenase (Cell Signaling Technology, Inc.) at 1:1000 dilution. All primary antibodies were detected using an horseradish peroxidase-linked anti-rabbit IgG antibody (Cell Signaling Technology) and visualised using a Luminescent Image Analyzer LAS-3000 (Fuji Photo Film Co., Ltd). Visualised bands were analysed by densitometry using Multi Gauge version 3.0 (Fuji Photo Film Co.).
Statistical analysis
Statistical differences between the three groups for all data except those of DNA microarray analyses were assessed by ANOVA with the Tukey–Kramer test, with P<0·05 considered indicative of significance. Real-time quantitative RT-PCR analysis was performed for the two groups (10C, 6C+4CP) similar to DNA microarray analysis, and the data were compared with Student’s t test.
Results
Dietary intake and growth
Body weight gain, water intake and food intake data are shown in Table 2. Food intake was significantly higher in the 6C+4CP group compared with the 14C and 10C groups. However, there were no significant differences in body weight gain between groups.
Table 2 Body weight and food and water intakes (Numbers; mean values with their standard errors)
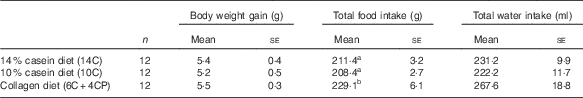
a,b Mean values with unlike superscript letters were significantly different (P<0·05).
Biochemical analysis of blood
Biochemical analysis of blood revealed that adiponectin and total, free and esterified cholesterol levels were significantly lower in the 6C+4CP group than in the 10C group (Fig. 1(A–D)). No significant changes were observed in other parameters including ALB, AST, ALT and TAG (data not shown). These results suggest that ingestion of CP lowers blood cholesterol levels without causing liver damages.

Fig. 1 Blood biochemical parameters. Plasma concentrations of total cholesterol (A), free cholesterol (B), esterified cholesterol (C) and adiponectin (D) are shown. Values are means (n 12 for each group), with standard errors represented by vertical bars. 14C, AIN-93M containing 14 % protein (as casein); 10C, AIN-93M-based low-protein diet containing 10 % protein (as casein); 6C+4CP, AIN-93M-based low-protein diet containing 6 % protein (as casein) plus 4 % protein (as collagen peptide). a,bMean values with unlike letters were significantly different (P<0·05).
Liver lipid content
No significant differences were detected in TAG or total and free cholesterol levels in the liver (data not shown).
Increased expressions of genes involved in lipid metabolism in the liver
The ingestion of CP resulted in the significant up-regulation of 485 probe sets. Enrichment analysis using GO annotation revealed that the expressions of genes related to fatty acid metabolism, cofactor metabolic processes, coenzyme metabolic processes and acyl-CoA metabolic processes were significantly up-regulated in the 6C+4CP group compared with the 10C group (Table 3). Enrichment analysis of the up-regulated gene set using KEGG annotation indicated that expressions of genes involved in biosynthesis of unsaturated fatty acids, PPAR signalling pathway and fatty acid metabolism were significantly up-regulated in the 6C+4CP group (Table 5). Notably, PPARα, one of the subtypes of PPAR, was up-regulated by CP ingestion. The significantly up-regulated genes related to the PPARα signalling pathway are summarised in Fig. 2(a). Among the up-regulated PPARα target genes involved in lipid metabolism were genes related to lipogenesis malic enzyme 1, NADP+-dependent, cytosolic (Me1), stearoyl-CoA desaturase 1 (Scd1), fatty acid desaturase 2 (Fads2), fatty acid β-oxidation (Acox1, carnitine palmitoyltransferase 1a, liver (Cpt1a), enoyl-CoA, hydratase/3-hydroxyacyl CoA dehydrogenase (Ehhadh), acetyl-CoA acyltransferase 1A (Acaa1a), cytochrome P450, family 4, subfamily a, polypeptide 10 (Cyp4a10), Cyp4a14) and fatty acid transport acyl-CoA synthetase long-chain family member 1 (Acsl1).

Fig. 2 Two representative biological processes modulated by collagen peptide ingestion. Up-regulated genes related to lipid metabolism, identified by Kyoto Encyclopedia of Genes and Genomes pathway analysis (a), and down-regulated genes related to the unfolded protein response (UPR), identified by Gene Ontology analysis (b) are summarised. IRE1, inositol-requiring enzyme 1; ER, endoplasmic reticulum.
Table 3 A list of biological process Gene Ontology (GO) terms and their component genes significantly up-regulated (false-discovery rate-corrected P-value<0·05) by collagen peptide ingestion

Decreased expression of genes involved in the unfolded protein response
The ingestion of CP resulted in the significant down-regulation of 395 probe sets. Enrichment analysis using GO annotation revealed that the expressions of genes related to steroid metabolic process, response to unfolded protein and response to protein stimulus were down-regulated by CP ingestion (Table 4). In contrast, no significantly enriched KEGG pathways were found in the down-regulated gene set. Down-regulated genes included in the ‘response to unfolded protein’ GO term are summarised in Fig. 2(b). In the 6C+4CP group, expressions of the gene encoding IRE1 (also known as Ern1; endoplasmic reticulum to nucleus signaling 1) and its target genes related to the degradation of unfolded proteins Der1-like domain family, member 2 (Derl2), Edem1, synovial apoptosis inhibitor 1, synoviolin (Syvn1), chaperon functions DnaJ (Hsp40) homolog, subfamily C, member 3 (Dnajc3) and endoplasmic reticulum (ER) stress-induced cell death mesencephalic astrocyte-derived neurotrophic factor (Manf) were down-regulated.
Table 4 A list of biological process Gene Ontology (GO) terms and their component genes significantly down-regulated (false-discovery rate-corrected P-value<0·05) by collagen peptide ingestion

Changes in inositol-requiring enzyme 1 and phospho-inositol-requiring enzyme 1 protein expression levels
The DNA microarray analysis identified two factors (PPARα and IRE1) as targets of CP ingestion. We performed Western blotting analysis of IRE1 protein because we did not expect a significant change in the unfolded protein response (UPR) following CP ingestion. As shown in Fig. 3(B), the level of IRE1 was significantly lower in the 6C+4CP group compared with the 14C group, whereas there was no significant difference compared with the 10C group. However, the level of the active form of IRE1 (p-IRE1) was significantly lower in the 6C+4CP group compared with the other two groups (Fig. 3(C)). The ratio of p-IRE1:IRE1 was also the lowest in the 6C+4CP group (Fig. 3(D)). These results suggest that ingestion of CP suppresses expression of the IRE1 gene and that the level of its active product (p-IRE1) declines in the liver following CP ingestion.

Fig. 3 Western blotting of inositol-requiring enzyme 1 (IRE1) and phosphor-IRE1 (p-IRE1). Western blotting images (A), quantitative data for IRE1 (B) and p-IRE1 (C) protein and p-IRE1:IRE1 ratio (D) are shown. The data were obtained from four replicate experiments and are represented as means with their standard errors (n 3 for each group). 14C, AIN-93M containing 14 % protein (as casein); 10C, AIN-93M-based low-protein diet containing 10 % protein (as casein); 6C+4CP, AIN-93M-based low-protein diet containing 6 % protein (as casein) plus 4 % protein (as collagen peptide). The vertical axis (B, C) shows the levels of IRE1 or p-IRE1 protein relative to the level of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) protein. a,bMean values with unlike letters were significantly different (P<0·05).
Discussion
In this study, food intake in the 6C+4CP group increased significantly compared with the 14C and 10C groups, indicating that energy intake was greater in the 6C+4CP group than in the other groups. However, there was no significant difference in body weight gain between the three groups. One explanation for this observation is that CP ingestion is associated with enhanced energy generation, as DNA microarray analysis of the liver revealed that the expression of genes related to fatty acid β-oxidation were up-regulated in the 6C+4CP group compared with the 10C group. Incidentally, the ratio of amino acids intake in the 6C+4CP group differed from those in the other two casein groups, owing to the difference in amino acid composition of each protein (casein, CP). However, no significant differences were observed in terms of body weight gain and liver lipid contents between the three groups. In addition, no significant changes in blood biomarkers indicative of pathological conditions were observed in any of the groups. These data suggest that the diet conditions examined this study are not associated with any severe adverse effects attributable to amino acid imbalance, such as growth retardation.
PPARα is a nuclear receptor that functions as a transcription factor, regulating the expression of genes involved in fatty acid catabolism( Reference Pyper, Viswakarma and Yu 24 – Reference Pawlak, Lefebvre and Staels 26 ). In the present study, the expression of PPARα and its target genes related to lipogenesis, fatty acid β-oxidation and fatty acid transport increased significantly after CP ingestion. This suggests the possibility that CP ingestion enhances lipid metabolism in the liver through activation of PPARα signalling, as reported for other natural products( Reference Mezei, Banz and Steger 27 – Reference Tan, Gao and Tao 31 ). Moreover, expression of the gene encoding Sirt1, which interacts with and activates PPARα signalling( Reference Purushotham, Schug and Xu 32 , Reference Moreno, Lombardi and Silvestri 33 ), was up-regulated in the 6C+4CP group (rank: 233 of the 485 significantly up-regulated probe sets, online Supplementary Table S1).
The expressions of genes involved in the biosynthesis of unsaturated fatty acids were also up-regulated in the 6C+4CP group, including PPARα targets such as Acaa1a, Acox1, Scd1 and Fads2 (Table 5). Unsaturated fatty acids conjugated with coenzyme A (i.e. fatty acyl-CoA) are transported into the peroxisome or the mitochondria and metabolised via the β-oxidation pathway( Reference Yan, Yang and Liu 34 ). As unsaturated fatty acids are also known to be endogenous ligands for PPARs( Reference Pyper, Viswakarma and Yu 24 – Reference Pawlak, Lefebvre and Staels 26 , Reference Forman, Chen and Evans 35 ), enhancement of this biosynthetic process is predicted to modulate PPARα signalling and β-oxidation.
Table 5 A list of Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways and their component genes significantly up-regulated (false-discovery rate-corrected P-value<0·05) by collagen peptide ingestion

Biochemical analysis of blood samples revealed that total, free and esterified cholesterol levels decreased significantly following CP ingestion in comparison with casein ingestion (Fig. 1(A–C)). This is consistent with previous studies that examined the lipid-lowering effects of collagen ingestion in humans and animals( Reference Wu, Fujioka and Sugimoto 3 , Reference Ratnayake, Sarwar and Laffey 11 – Reference Terpstra, Hermus and West 15 ). In the present study, significant differences were observed neither in the level of blood TAG nor in liver lipid contents, although our DNA microarray data suggest that liver lipid metabolism is enhanced by CP ingestion. Similar results that reduction in blood cholesterol without changes in TAG were also observed in our previous study in rats( Reference Koyama and Kusubata 14 ). On the other hand, there are several reports that serum TAG is reduced by CP ingestion( Reference Ratnayake, Sarwar and Laffey 11 , Reference Oliveira, Portugal and Cara 13 ), and one explanation for the reduction is described as increase in faecal excretion( Reference Oliveira, Portugal and Cara 13 ). This discrepancy can be partly accounted for by the differences in the experimental conditions or timing of sampling, but the precise reason is still unclear. However, it is certain that CP ingestion affects lipid metabolism, and we conceive that PPARα signalling may be involved in the mechanism of the CP effect. In the 6C+4CP group, the expressions of genes related to primary bile acid biosynthesis were down-regulated (Table 4; steroid metabolic process). It is known that the process of primary bile acid biosynthesis is the major mechanism for the removal of cholesterol from the body, as by synthesising primary bile acid via oxidation of cholesterol. Although PPARα signalling was up-regulated in the 6C+4CP group (Tables 3 and 5), certain genes of PPARα targets (Cyp7a1, Cyp8b1, Cyp27a1 and Acox2; related to primary bile acid biosynthesis) were down-regulated. Our data suggest that the decrease in blood cholesterol levels associated with CP ingestion is not solely due to enhancement of primary bile acid biosynthesis. We presumed that the down-regulation of these genes occurred as a reaction of suppressing excess decrease of cholesterol for cholesterol homoeostasis in the body. It has been reported that some proteins other than collagen, such as soyabeans and whey, also have cholesterol-lowering effects, and the main mechanism of the effects is the suppression of cholesterol absorption (i.e. faecal excretion of cholesterol)( Reference Nagaoka, Awano and Nagata 36 , Reference Nagaoka, Futamura and Miwa 37 ). We presumed that CP ingestion caused the continuous decrease in internal cholesterol, thus the process of primary bile acid biosynthesis was suppressed. In addition, the bile acids are secreted into the intestine and recirculated to the liver by the enterohepatic circulation; there is a negative-feedback mechanism mediated by the bile acids themselves in the liver, suppressing the expression of the genes related to primary bile acid biosynthesis including Cyp7a1 and Cyp8b1 ( Reference Wang, Lee and Bundman 38 ). It is possible that this mechanism may be related to our results in this study. To clarify the precise mechanisms involved in the cholesterol-lowering effects of CP ingestion, more detailed investigations should be performed that focus on the absorption, excretion, synthesis and transport of cholesterol in each organ. However, it is a new finding of our study that the effect of blood cholesterol lowering by CP ingestion is not due to enhancement of primary bile acid biosynthesis. In addition, the level of adiponectin in the blood was significantly lower in the 6C+4CP group compared with the other groups. Decreased blood adiponectin levels are associated with the development of various pathological conditions( Reference Tan, Gao and Tao 31 , Reference Matsuzawa, Funahashi and Kihara 39 , Reference Fisman and Tenenbaum 40 ). However, adiponectin levels in the present study were within the normal range for mice (17·0–77·0 mg/l), and no obesity or increase in blood/liver TAG was observed in the 6C+4CP group. Thus, we conclude that the reduction of blood adiponectin level by CP ingestion does not lead to obesity.
During protein synthesis, unfolded proteins can accumulate in the ER and generate cellular stress, and the UPR system limits the effects of this type of cellular stress( Reference Ron and Walter 41 , Reference Jo, Choe and Shin 42 ). In the present study, the expressions of IRE1 and its target genes were down-regulated in the 6C+4CP group in comparison with the 10C group (Table 4). p-IRE1 functions as an RNase, splicing the mRNA of X-box-binding protein (Xbp1), a key UPR transcription factor, to produce its active form. The genes induced by ER-stress sensing by IRE1 include those encoding proteases and chaperones, which function to reduce levels of accumulated proteins( Reference Kaneko, Ishiguro and Niinuma 43 – Reference Oda, Okada and Yoshida 45 ). A direct search of the microarray data revealed that expression of the Xbp1 gene was also down-regulated in the 6C+4CP group (rank: 149, 271 of the 395 significantly down-regulated probe sets, online Supplementary Table S1). Taken together, these results suggest that ER stress is suppressed by CP ingestion, although the possibility of general reduction in protein synthesis requires further examination. Collagen contains the unique amino acid hydroxyproline (Hyp), and it was reported that ingested CP is digested and absorbed in part as Hyp-containing oligopeptides, such as prolylhydroxyproline (Pro-Hyp) or hydroxyprolylglycine (Hyp-Gly)( Reference Iwai, Hasegawa and Taguchi 46 – Reference Shigemura, Akaba and Kawashima 49 ). The blood concentration of Pro-Hyp can reach approximately 50 μm by 1–2 h after ingestion of 25 g of CP( Reference Taga, Kusubata and Ogawa-Goto 50 ). Pro-Hyp exhibits physiological activity against cultured skin fibroblasts( Reference Shigemura, Iwai and Morimatsu 51 ) and pre-adipocytes( Reference Minaguchi, Tometsuka and Koyama 52 ), whereas Hyp-Gly modulates the differentiation of cartilage cells in vitro and in vivo ( Reference Nakatani, Mano and Sampei 53 ). These data suggest that collagen-derived oligopeptides are responsible, at least in part, for the beneficial in vivo effects of collagen ingestion. Although the precise mechanisms underlying the modulation of cell function by collagen oligopeptides are currently unclear, it seems possible that they are incorporated into cells through the proton-coupled peptide transporters 1 and 2 (PEPT1 and PEPT2) and peptide/histidine transporters 1 and 2 (PHT1 and PHT2 or Ci1 in mice), the genes of which are expressed in Pro-Hyp-responsive cells( Reference Minaguchi, Tometsuka and Koyama 52 ). In this context, it is noteworthy that proton-coupled peptide transporter genes are expressed in the liver( Reference Smith, Clémençon and Hediger 54 ).
In conclusion, the results of the present study focusing on the effects of collagen ingestion on liver gene expression reveal that ingested CP alters the expressions of genes related to lipid metabolism. Moreover, the blood cholesterol-lowering effect of CP ingestion was reproduced in the present study, and it was presumed that the effect is not due to enhancement of primary bile acid biosynthesis according to DNA microarray analysis of the liver. Although the precise mechanisms remain unclear, novel functions of collagen in the broader metabolic pathways of the liver are predicted on the basis of the decreased expression of genes related to the UPR.
Acknowledgements
The authors are grateful to Professor Hisanori Kato and Dr Lila Otani (The University of Tokyo) for meaningful discussions regarding this study.
This research received no specific grant from any funding agency or from commercial or not-for-profit sectors.
Conceived and designed the experiments: C. T., Y.-i. K., K. T., K. A. and Y. N. Performed the experiments: C. T., Y.-i. K., T. I., T. T., M. T. and K. T. Analysed the data: C. T. and Y. N. Wrote the paper: C. T., Y.-i. K. and Y. N.
The authors declare that there are no conflicts of interest.
Supplementary material
For supplementary material/s referred to in this article, please visit https://doi.org/10.1017/S0007114516004384











