PUFA are a group of critical nutrients that modulate brain development, cognition and several diseases, including CVD, cancers and diabetes(Reference Simopoulos1–Reference Elajami, Alfaddagh and Lakshminarayan4). PUFA are classified as n-3 and n-6 fatty acids (FA).
Humans derive long-chain (LC) PUFA directly from their diet and can synthesise them endogenously from their essential n-6 and n-3 precursors, linoleic acid (LA) and α-linolenic acid (ALA), respectively(Reference Yeates, Love and Engstrom5). This process requires a consecutive series of desaturation involving Δ-5 (D5D) and Δ-6 fatty acid desaturases (D6D) encoded by the fatty acid desaturase 1 (FADS1) and fatty acid desaturase 2 (FADS2) genes, respectively, and elongation reactions. Human desaturase complementary DNA was first cloned by Cho et al. (Reference Cho, Nakamura and Clarke6,Reference Cho, Nakamura and Clarke7) and later identified in a cluster on chromosome 11 (11q12–13.1)(Reference Marquardt, Stohr and White8). D5D and D6D are expressed across several tissues but predominantly expressed in the liver(Reference Cho, Nakamura and Clarke6,Reference Cho, Nakamura and Clarke7) . LA and ALA are metabolised by the same series of enzymes, and EPA and DHA are produced at limited conversion rates of 0·2–6 % and less than 0·05 %, respectively, in men(Reference Burdge9). The synthesis efficiency of endogenous EPA and DHA is thought to be affected by gene polymorphisms. A 5-locus haplotype explains 1·4, 5·2 and 27·7 % of the variability in DHA, EPA and arachidonic acid (AA) levels, respectively(Reference Minihane10).
As described, genetic variation in FADS appears to be important for modulating the LC-PUFA status. SNP in FADS1 may affect LC-PUFA production and consequently alter FA levels(Reference Harslof, Larsen and Ritz11). The rs174547 SNP is located in intron 9 of FADS1 (Reference Kim, Kim and Yoo12). Over the past two decades, several genome-wide association studies have reported the associations of rs174547 with FA(Reference Dorajoo, Sun and Han13–Reference Hu, Li and Lu15). Previously, we demonstrated that SNP of the rs174547 (T/C) genotype in FADS1 were associated with the FA composition in Chinese populations(Reference Ding, Liu and Li16). rs174547 is a functional variant associated with decreased FADS1 expression in the human liver(Reference Takkunen, de Mello and Schwab17). Several studies(Reference Kim, Kim and Yoo12,Reference Ding, Liu and Li16,Reference Merino, Johnston and Clarke18–Reference Nita, Kawabata and Kagawa26) have focused on this variant and its association with PUFA levels. Among the tag SNP of FADS1 gene, rs174547 tags up to seven other SNP in an 11-kb genomic region of FADS1 with a linkage disequilibrium threshold of R 2 > 0·8(Reference Ding, Liu and Li16).
In the present study, we performed a meta-analysis to determine the effects of rs174547 in FADS1 on PUFA levels.
Methods
Literature retrieval
Four databases (Pubmed, Web of Science, China National Knowledge Infrastructure (CNKI) and Wanfang databases) were searched to retrieve related literature with key words such as fatty acid, SNP, FADS1 gene and rs174547 published in English and Chinese Language before 5 October 2020. The detailed search strategy is presented in Supplementary Table 1.
Assessment of eligibility
Studies were suggested to be eligible if they meet the following inclusion criteria: (1) studies reported in Chinese or English; and (2) full-text applicable, with access to required materials and data from the authors and (3) studies with good design quality were selected for analysis. Studies were excluded if they were (1) duplicated publications, (2) abstracts, case reports/series, comments, editorial articles, summary, animal/plant/cell studies, reviews or meta-analysis, (3) data expressed as medians (25–75th percentiles) and (4) missing data that were not presented in the literature and we did not get reply from the authors who published those studies.
Data extraction and quality assessment
Author, publication year, country, age, sample size, genotypes, measurement method, tissues and FA content were extracted from the included studies. Two investigators independently extracted data from included literature, and any disagreement was resolved by discussion. The Newcastle–Ottawa Scale(Reference Wang, Yang and Wei27) was used to assess the quality of included study, and quality assessment results were presented in Supplementary Table 2. A study that scored ≥6 points (total is 8 points) was defined as high quality. The three genotypes (TT, TC and CC) of subjects were divided into major homozygotes carriers and minor allele carriers groups (TT and TC + CC) using the method of weighing according to retrieved studies(Reference Chen, Wu and Zhang28).
Statistical analysis
The meta-analysis of data was performed using Stata12.0 software. Heterogeneity of the included studies was performed by Q test and I 2 test. When P values was >0·1 and I 2 values was ≤50 %, a fixed effects model was used; otherwise, a random effects model was used. The random effect model was selected and effect indexes of each study were calculated as standardised mean difference (SMD) and 95 % CI. Heterogeneity of studies was assessed by subgroup analysis and sensitivity analysis. Subgroup analysis was performed by region, tissue and score of quality of studies group. Metafunnel and Egger’s or Begger’s test were used to assess publication bias. P < 0·05 was considered to be statistically significant.
Results
Included literature
Total of 1967 relevant publications were searched. Based on the inclusion and exclusion criteria, eleven high-quality publications were finally included in the meta-analysis; the flow diagram of retrieval process was shown in Fig. 1. A Population; Exposure; Comparision and Outcomes (PECO) table for included papers is presented in Supplementary Table 3.

Fig. 1. Flow diagram of the selection process of the literature search.
Study characteristics
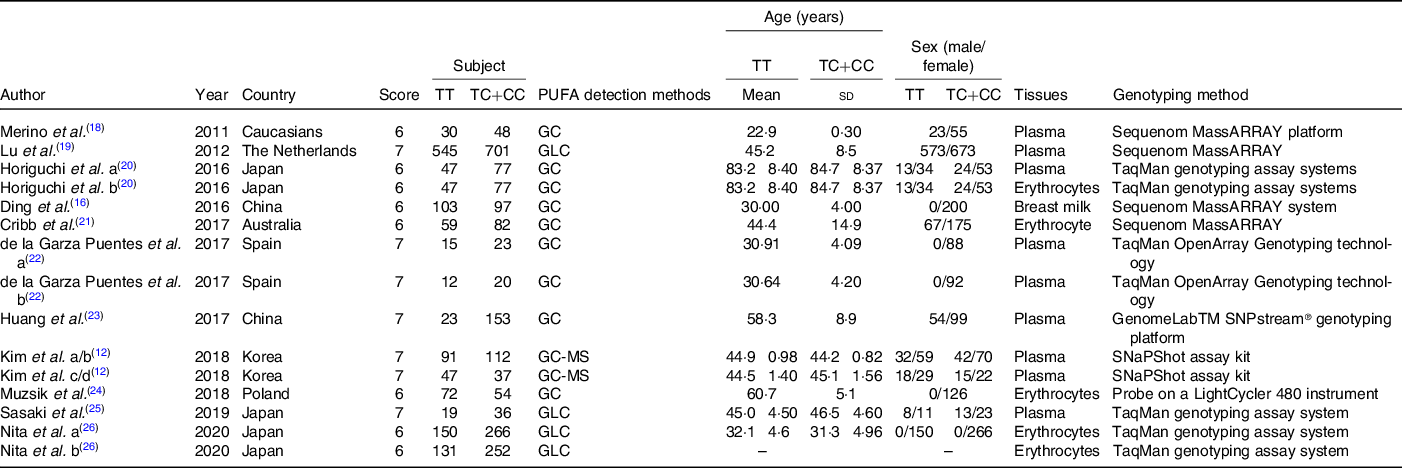
Data from seventeen trials were extracted from eleven literature (since one literature focused on the study of PUFA in plasma and erythrocytes at the same time, one focused on the study of PUFA in mothers and fetuses, one has two cohorts study in normal-weight and overweight population at two different times, and another one has two cohorts study in normal-weight and overweight population). A total of 3713 individuals (1529 TT and 2184 TC + CC) were included. Subjects of these studies were from Asia, Europe and Oceania. These studies detected the FA composition of plasma, erythrocyte and breast milk. The Newcastle–Ottawa Scale scores of the studies varied from 6 to 7 points, indicating the good quality of the included studies. More information or characteristics of the included studies are summarised in Table 1.
Table 1. Characteristics of studies included in the meta-analysis
(Mean values and standard deviations)
Results of meta-analysis
Effects of rs174547 in FADS1 on linoleic acid levels
The effects of rs174547 on LA levels are shown in Fig. 2(a). The LA level in minor C allele carriers was significantly higher than in the TT genotype group (SMD: 1·16, 95 % CI 0·65, 1·68, P < 0·001), and significant heterogeneity (I 2 = 96·5 %, P < 0·001) was observed. Subgroup analysis demonstrated that the LA level in minor C allele carriers was significantly higher than that in the TT genotype group across Asian populations, Oceanian populations, plasma samples, erythrocyte samples and study quality groups (score = 7 and score = 6), respectively (Table 2).
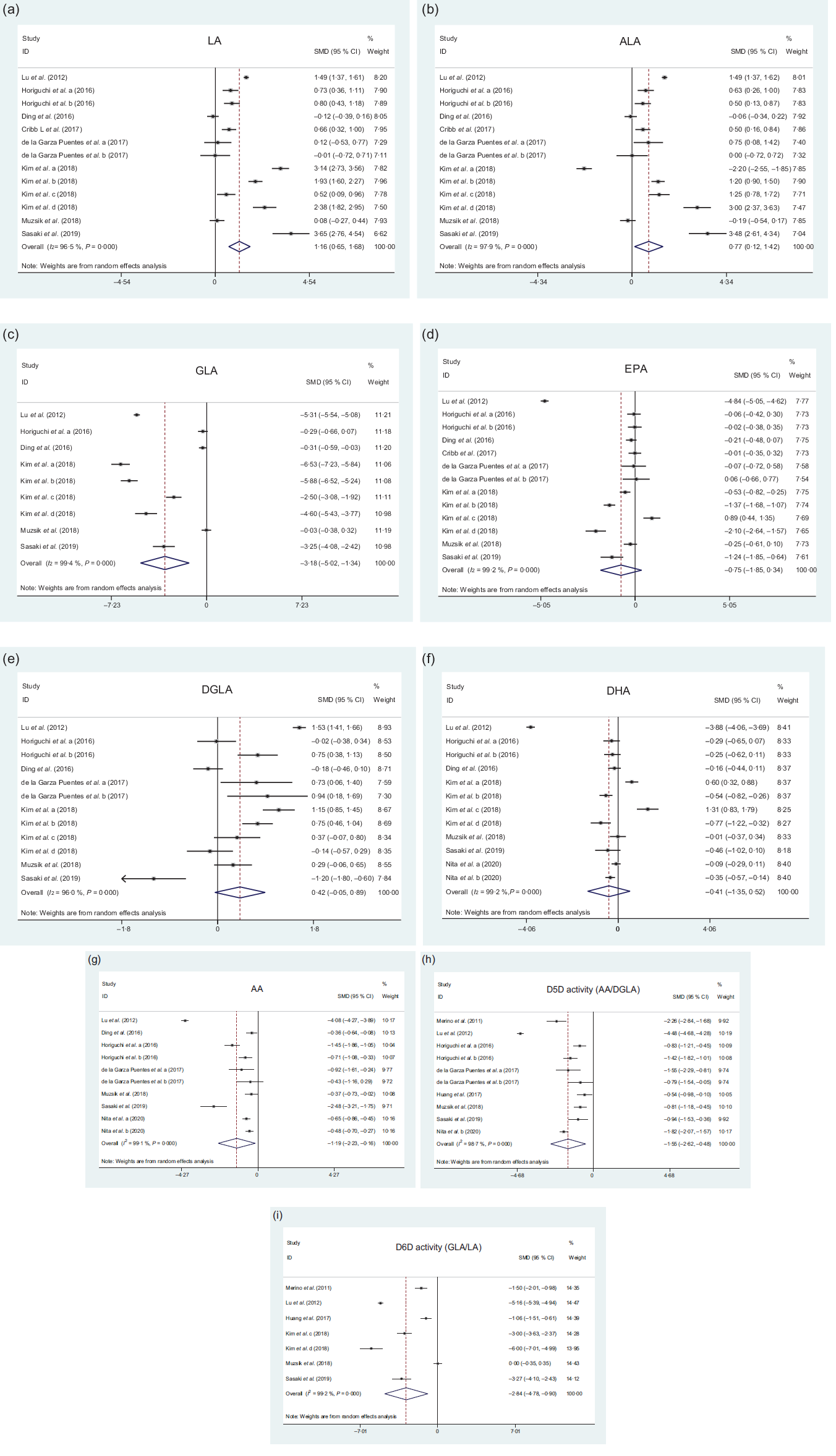
Fig. 2. Forest plots showing PUFA levels difference between minor C allele carriers and TT genotype of rs174547 in FADS1. (a) Forest plot of linoleic acid (LA); (b) forest plot of α-linolenic acid (ALA); (c) forest plot of γ-linolenic acid (GLA); (d) forest plot of EPA; (e) forest plot of dihomo-γ-linolenic acid (DGLA); (f) forest plot of DHA; (g) forest plot of arachidonic acid (AA); (h) forest plot of Δ-5 desaturase (D5D) activity; (i) forest plot of Δ-6 desaturase (D6D) activity. D5D and D6D activities were assessed by the ratio of AA to DGLA and GLA to LA, respectively.
Table 2. Subgroup meta-analysis of the SNP rs174547 in FADS1 on the level of long-chain-PUFA (Numbers, standardised mean differences (SMD) and 95 % confidence intervals)
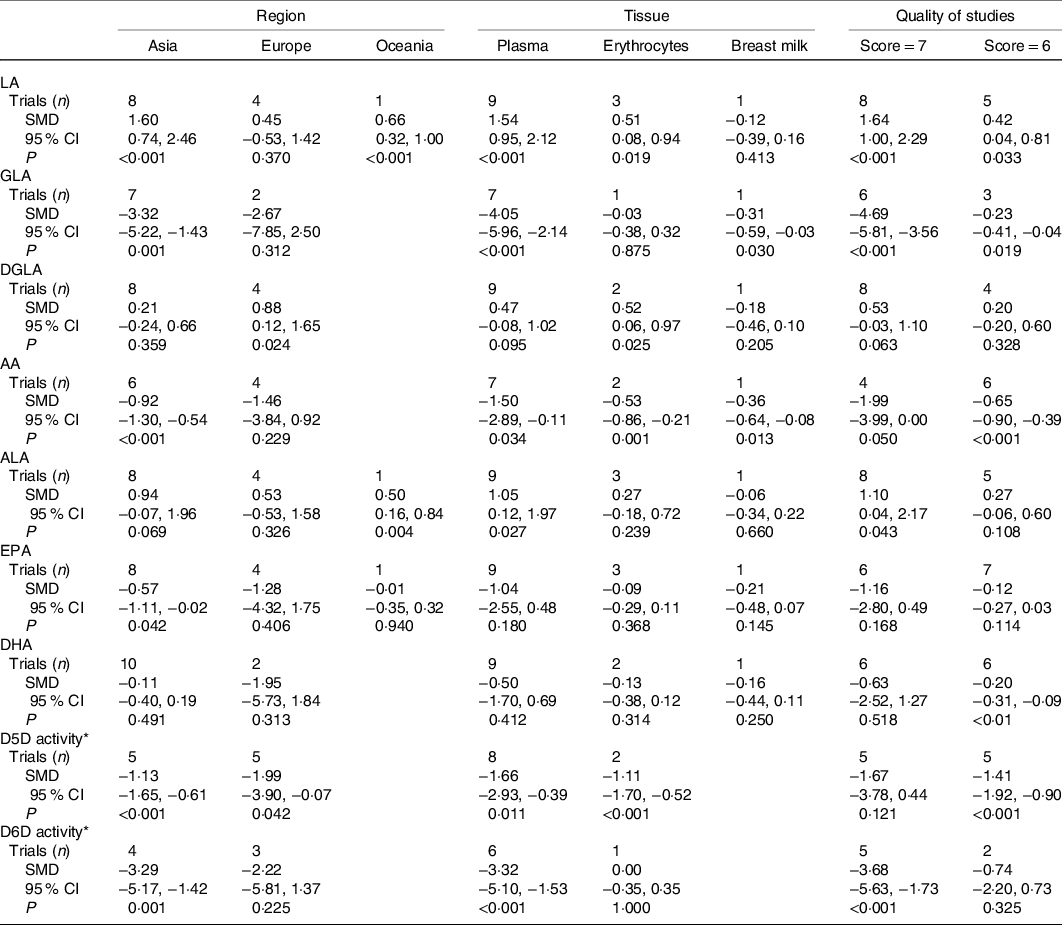
LA, linoleic acid; GLA, γ-linolenic acid; DGLA, dihomo-γ-linolenic acid; AA, arachidonic acid; ALA, α-linolenic acid; D5D, Δ-5 desaturase; D6D, Δ-6 desaturase.
* D5D and D6D activities were assessed by the ratio of AA to DGLA and GLA to LA, respectively.
Effects of rs174547 in FADS1 on γ-linolenic acid level
The effects of rs174547 on γ-linolenic acid (GLA) level are shown in Fig. 2(c). The GLA level in minor C allele carriers was significantly lower than that in the TT genotype group (SMD: −3·18, 95 % CI −5·02, −1·34, P = 0·001), and significant heterogeneity (I 2 = 99·4 %, P < 0·001) was observed. Subgroup analysis demonstrated that the GLA level in minor C allele carriers was significantly lower than that in the TT genotype group across Asian populations, plasma samples, breast milk samples and study quality groups (score = 7 and score = 6), respectively (Table 2).
Effects of rs174547 in FADS1 on dihomo-γ-linolenic acid level
The effects of rs174547 on dihomo-γ-linolenic acid (DGLA) level are shown in Fig. 2(e). The difference of DGLA level was not significant between minor C allele carriers and the TT genotype group (SMD: 0·42, 95 % CI −0·05, 0·89, P = 0·079), and significant heterogeneity (I 2 = 96·0 %, P < 0·001) was observed. Subgroup analysis demonstrated that the DGLA level in minor C allele carriers was significantly higher than that in the TT genotype group in European populations and erythrocyte samples group (Table 2).
Effects of rs174547 in FADS1 on arachidonic acid level
The effects of rs174547 on AA level are shown in Fig. 2(g). The AA level in minor C allele carriers was significantly lower than that in the TT genotype group (SMD: −1·19, 95 % CI −2·23, −0·16, P = 0·024), and significant heterogeneity (I 2 = 99·1 %, P < 0·001) was observed. Subgroup analysis demonstrated that the AA level in minor C allele carriers was significantly higher than that in the TT genotype group across Asian populations, all tissue groups and all study quality groups, respectively (Table 2).
Effects of rs174547 in FADS1 on α-linolenic acid level
The effects of rs174547 on ALA level are shown in Fig. 2(b). The ALA level in minor C allele carriers was significantly higher than that in the TT genotype group (SMD: 0·77, 95 % CI 0·12, 1·42, P = 0·020), and significant heterogeneity (I 2 = 97·9 %, P < 0·001) was observed. Subgroup analysis demonstrated that the ALA level in minor C allele carriers was significantly higher than that in the TT genotype group in Oceanian populations, plasma samples and study quality group (score = 7) (Table 2).
Effects of rs174547 in FADS1 on EPA level
The effects of rs174547 on EPA level are shown in Fig. 2(d). The difference of EPA level was not significant between minor C allele carriers and the TT genotype group (SMD: −0·75, 95 % CI −1·85, 0·34, P = 0·177), and significant heterogeneity (I 2 = 99·2 %, P < 0·001) was observed. Subgroup analysis demonstrated that the EPA level in minor C allele carriers was significantly lower than that in the TT genotype group in Asian populations group (Table 2).
Effects of rs174547 in FADS1 on DHA level
The effects of rs174547 on DHA level are shown in Fig. 2(f). The difference of DHA level was not significant between minor C allele carriers and the TT genotype group (SMD: −0·41, 95 % CI −1·35, 0·52, P = 0·388), and significant heterogeneity (I 2 = 99·2 %, P < 0·001) was observed. Subgroup analysis demonstrated that the DHA level in minor C allele carriers was significantly lower than that in the TT genotype group in study quality group (score = 6) (Table 2).
Effects of rs174547 in FADS1 on Δ-5 fatty acid desaturase activity
The effects of rs174547 on D5D activity (the ratio of AA to DGLA) are shown in Fig. 2(h). The D5D activity in minor C allele carriers was significantly lower than that in the TT genotype group (SMD: −1·55, 95 % CI −2·62, −0·48, P = 0·005), and significant heterogeneity (I 2 = 98·7 %, P < 0·001) was observed. Subgroup analysis demonstrated that the D5D activity in minor C allele carriers was significantly lower than that in the TT genotype group across Asia populations, European populations, plasma samples, erythrocyte samples and quality of studies (score = 6) group (Table 2).
Effects of rs174547 in FADS1 on Δ-6 fatty acid desaturase activity
The effects of rs174547 on D6D activity (the ratio of GLA to LA) are shown in Fig. 2(i). The D6D activity in minor C allele carriers was significantly lower than that in the TT genotype group (SMD: −2·84, 95 % CI −4·78, −0·90, P = 0·004), and significant heterogeneity (I 2 = 99·2 %, P < 0·001) was observed. Subgroup analysis demonstrated that the D6D activity in minor C allele carriers was significantly lower than that in the TT genotype group across Asian populations, plasma populations and study quality group (score = 7) (Table 2).
Sensitivity analysis
Sensitivity analysis was performed by removing studies one by one to evaluate the stability of the results. As shown in Supplementary Fig. 1, although each study was successively removed, the results did not alter obviously in LA, GLA, D5D and D6D activities, indicating the high stability of the meta-analysis results. When the study from Sasaki et al. (Reference Sasaki, Sueyasu and Tokuda25) was excluded, the difference of ALA (SMD: 0·57, 95 % CI −0·09, 1·22, P = 0·092) between minor C allele carriers and the TT genotype group and AA (SMD: −1·06, 95 % CI −2·16, 0·05, P = 0·060) levels was not statistically significant, and DGLA (SMD: 0·56, 95 % CI 0·10, 1·01, P = 0·016) level in minor C allele carriers was significantly higher than that in the TT genotype group. When the study from Lu et al. (Reference Lu, Vaarhorst and Merry19) was excluded, the EPA (SMD: −0·41, 95 % CI −0·80, −0·01, P = 0·043) level in minor C allele carriers was significantly lower than that in the TT genotype group.
Publication bias
The funnel plots of the SNP rs174547 on LC-PUFA level did not reveal substantial publication bias (Supplementary Fig. 2), indicating no significant publication bias of results. Egger’s or Begger’s test was carried out to analyse the publication bias. The analysis results showed the presence of publication bias in DGLA (Egger’s test P = 0·006) and D5D activity (Egger’s test P = 0·041) (Supplementary Table 4).
Discussion
In the present study, the associations between rs174547 in FADS1 and seven types of FA, D5D activity and D6D activity were assessed based on the pooled results from eleven papers(Reference Kim, Kim and Yoo12,Reference Ding, Liu and Li16,Reference Merino, Johnston and Clarke18–Reference Nita, Kawabata and Kagawa26) . The results demonstrated that minor C allele carriers of rs174547 had higher LA and ALA levels, lower GLA and AA levels, and lower D5D and D6D activities than the TT genotype group. Desaturation of LC-PUFA, derived from ALA and LA through elongation and desaturation, is regulated by D5D and D6D(Reference Juan, Huang and Jiang29). We observed a weaker association between LA and GLA among carriers of the minor C allele of a representative SNP in FADS1 (rs174547), suggesting a lower rate of LA-to-GLA conversion in these individuals. The increased proportions of substrates (LA) and decreased products (GLA) demonstrated lower D6D activity in the n-6 LC-PUFA synthetic pathway. Additionally, minor C allele carriers of the SNP rs174547 (FADS1) were associated with decreased D5D and D6D activities. This suggests that FADS1 variants influence the rate of conversion of LA into other n-6 PUFA, indicating an associated loss-of-function effect. GLA is generated from LA catalysed by D6D. However, the GLA level and D6D activity are thought to be associated with genetic polymorphisms in FADS1 (Reference Kim, Kim and Yoo12), consistent with our findings. The SNP rs174547 is linked with the SNP rs174570 in FADS2, which is related to D6D, as they are in the same linkage disequilibrium block(Reference Nakayama, Bayasgalan and Tazoe30) (r 2 = −1; HapMap JPT + CHB panel (Japanese and Han Chinese individuals)). Thus, the rs174547 genotype may reflect the entire metabolic process of n-6 PUFA conversion from LA to AA, including desaturation by D5D and D6D. Variants in FADS1 play important roles in regulating n-6 PUFA. Moreover, the SNP rs174547 was previously recognised for its association with n-6 PUFA and desaturase activities in the genome-wide association studies (Reference Guan, Steffen and Lemaitre14).
A meta-analysis of genome-wide association studies(Reference Lemaitre, Tanaka and Tang31) that reported the association of minor alleles of SNP in FADS1 with higher levels of ALA and lower levels of EPA and DPA. Although these results were not completely consistent with our findings, they indicate that the precursor and product FA compositions differed when there was a mutation in the FADS1 SNP. To date, there are few and conflicting studies on the effects of dietary PUFA and FADS1 interactions. The associations of FADS1 with the PUFA levels did not vary depending on the frequency of fatty fish consumption, suggesting that the genetic effects are independent of dietary intake consumption in the studied populations(Reference Juan, Huang and Jiang29). Other studies suggested that the associated effects of SNP in FADS1 on FA concentrations can be modified by dietary PUFA intake(Reference Takkunen, de Mello and Schwab17).
We performed subgroup analysis because of the marked heterogeneity observed in our analysis. We found that different ethnicities and tissue types may contribute to the observed heterogeneity. In stratification analysis, we found that minor C allele carriers of rs174547 had higher LA and lower GLA levels and lower D6D activities in the plasma samples and in Asian populations than the TT genotype group. This suggests that the rs174547 polymorphism is a crucial genetic factor in Asian populations. It may also be possible to predict plasma n-6 FA LA and GLA levels in Asian populations based on genotype. However, the number of other ethnic group studies in our meta-analysis was small; only one study in Oceania was included(Reference Cribb, Murphy and Froud21). Moreover, they focused on PUFA in the plasma and erythrocytes, with a single study focusing on breast milk.
Growing evidence has suggested that PUFA play important roles in disease. The n-3 and n-6 PUFA metabolic pathways participate in several inflammatory processes(Reference Saunders, Ramsden and Sherazy32). However, the interactions of n-3 and n-6 PUFA in inflammation are complex and poorly understood. A previous study(Reference Saunders, Ramsden and Sherazy32) proposed that increasing dietary intake of the n-6 PUFA AA or its precursor LA can increase inflammation. However, studies in healthy human adults have demonstrated that increased intake of AA or LA does not increase concentrations of inflammatory factors. Moreover, epidemiological studies suggested that AA and LA are associated with reduced inflammation(Reference Innes and Calder33). n-3 LC-PUFA have also been observed in a variety of inflammatory diseases, including asthma(Reference Best, Gold and Kennedy34), rheumatoid arthritis(Reference Goldberg and Katz35) and cancer(Reference Sakai, Kakutani and Horikawa36). Regular dietary intake of n-6 FA may help prevent and treat hypertension in a healthy population. In contrast, studies have also shown that regular intake of n-6 FA in people with diabetes may increase the risk of hypertension(Reference Nakamura, Hara and Tsujiguchi37). Recently, a negative correlation between n-3 FA and CVD was demonstrated(Reference Tortosa-Caparros, Navas-Carrillo and Marin38). The reciprocal associations support the important role of genetic variation in the pathway for circulating levels of PUFA in diseases.
Our meta-analysis had some limitations. First, our pooled results were based on raw data, with no adjustments made to accommodate for influencing factors. This was because of missing and incomplete information. Second, we did not investigate the interactions between gene and diet or gene and gene because of the limited access to raw data from eligible studies. To comprehensively analyse the effects of rs174547 on PUFA levels, we did not exclude observational studies(Reference Sasaki, Sueyasu and Tokuda25) or clinical trials(Reference Lu, Vaarhorst and Merry19), potentially creating selection bias. Thus, an increased number of original studies with comprehensive raw data are required to confirm the associations between FADS1 polymorphisms and PUFA levels. We focused on only one rs174547 variant within the FADS gene cluster and its association with PUFA levels. Some additional studies may have been missed because they used a different SNP marker to investigate comparable associations. Broader studies are needed to summarise the evidence of the genetic contribution of the FADS gene cluster to the PUFA level.
In conclusion, minor C allele carriers of the SNP rs174547 in FADS1 were associated with decreased activity of D5D and D6D. Stratification analysis revealed that minor C allele carriers of rs174547 were associated with decreased activity of D6D in plasma and Asian populations and decreased activity of D5D in the plasma and erythrocytes samples, as well as Asian and European populations.
Acknowledgements
The present study was supported by the Department of Science and Technology of Jilin Province in China (20180101122JC).
L. X. conceived the study design; Y. J. and W. X. searched and selected the trials, N. U., H. Y. and Y. Wu extracted, analysed and interpreted the data; Y. Wang and Y. T. drafted the manuscript.
The authors had no conflicts of interest relevant to this article to disclose.
Supplementary material
To view supplementary material for this article, please visit https://doi.org/10.1017/S0007114520005103







