Crossref Citations
This article has been cited by the following publications. This list is generated based on data provided by
Crossref.
Faria, Ricardo António Silva
Maiorano, Amanda Merchi
Bernardes, Priscila Arrigucci
Pereira, Guilherme Luis
Silva, Marina Gabriela Berchiol
Curi, Rogério Abdallah
and
Silva, Josineudson Augusto II Vasconcelos
2018.
Assessment of pedigree information in the Quarter Horse: Population, breeding and genetic diversity.
Livestock Science,
Vol. 214,
Issue. ,
p.
135.
Ablondi, Michela
Vasini, Matteo
Beretti, Valentino
Superchi, Paola
and
Sabbioni, Alberto
2018.
Exploring genetic diversity in an Italian horse native breed to develop strategies for preservation and management.
Journal of Animal Breeding and Genetics,
Vol. 135,
Issue. 6,
p.
450.
Bussiman, F.O.
Perez, B.C.
Ventura, R.V.
Peixoto, M.G.C.D.
Curi, R.A.
and
Balieiro, J.C.C.
2018.
Pedigree analysis and inbreeding effects over morphological traits in Campolina horse population.
Animal,
Vol. 12,
Issue. 11,
p.
2246.
Giontella, A.
Pieramati, C.
Silvestrelli, M.
and
Sarti, F.M.
2019.
Analysis of founders and performance test effects on an autochthonous horse population through pedigree analysis: structure, genetic variability and inbreeding.
Animal,
Vol. 13,
Issue. 1,
p.
15.
Baena, Marielle Moura
Gervásio, Izally Carvalho
Rocha, Renata de Fátima Bretanha
Procópio, Alessandro Moreira
de Moura, Raquel Silva
and
Meirelles, Sarah Laguna Conceição
2020.
Population structure and genetic diversity of Mangalarga Marchador horses.
Livestock Science,
Vol. 239,
Issue. ,
p.
104109.
Giontella, Andrea
Sarti, Francesca Maria
Cardinali, Irene
Giovannini, Samira
Cherchi, Raffaele
Lancioni, Hovirag
Silvestrelli, Maurizio
and
Pieramati, Camillo
2020.
Genetic Variability and Population Structure in the Sardinian Anglo-Arab Horse.
Animals,
Vol. 10,
Issue. 6,
p.
1018.
Plate, Manuel
Bernstein, Richard
Hoppe, Andreas
and
Bienefeld, Kaspar
2020.
Long-Term Evaluation of Breeding Scheme Alternatives for Endangered Honeybee Subspecies.
Insects,
Vol. 11,
Issue. 7,
p.
404.
Shelyov, A. V.
Kopylov, K. V.
Kramarenko, S. S.
and
Kramarenko, A. S.
2020.
Genetic structure of different equine breeds by microsatellite DNA loci.
Agricultural Science and Practice,
Vol. 7,
Issue. 2,
p.
3.
Marín Navas, Carmen
Delgado Bermejo, Juan Vicente
McLean, Amy Katherine
León Jurado, José Manuel
Rodriguez de la Borbolla y Ruiberriz de Torres, Antonio
and
Navas González, Francisco Javier
2021.
Discriminant Canonical Analysis of the Contribution of Spanish and Arabian Purebred Horses to the Genetic Diversity and Population Structure of Hispano-Arabian Horses.
Animals,
Vol. 11,
Issue. 2,
p.
269.
Yahagi Rodrigues, Larissa
da Silva Faria, Ricardo António
and
de Vasconcelos Silva, Josineudson Augusto II.
2021.
Analysis of the Pedigree and Ancestors of the Cutting Population of the Quarter Horse Breed.
Journal of Equine Veterinary Science,
Vol. 99,
Issue. ,
p.
103385.
Próchniak, Tomasz
Kasperek, Kornel
Knaga, Sebastian
Rozempolska-Rucińska, Iwona
Batkowska, Justyna
Drabik, Kamil
and
Ziȩba, Grzegorz
2021.
Pedigree Analysis of Warmblood Horses Participating in Competitions for Young Horses.
Frontiers in Genetics,
Vol. 12,
Issue. ,
Ivanković, Ante
Bittante, Giovanni
Konjačić, Miljenko
Kelava Ugarković, Nikolina
Pećina, Mateja
and
Ramljak, Jelena
2021.
Evaluation of the Conservation Status of the Croatian Posavina Horse Breed Based on Pedigree and Microsatellite Data.
Animals,
Vol. 11,
Issue. 7,
p.
2130.
Faria, Ricardo António Silva
Vicente, António Pedro Andrade
Ospina, Alejandra Maria Toro
and
Silva, Josineudson Augusto II Vasconcelos
2021.
Pedigree analysis of the racing line Quarter Horse: Genetic diversity and most influential ancestors.
Livestock Science,
Vol. 247,
Issue. ,
p.
104484.
Ferreira, J.L.
Ferraz, J.B.S.
Bussiman, F.O.
Rodrigues, M.R.
Bueno, R.S.
Sousa, L.A.
Carvalho, M.E.
Santos, H.D.
Toniolli, R.
Mello, S.Q.S.
and
Sousa, L.F.
2022.
Genetic variability of Guzerat cattle raised in northern Brazil, based on pedigree analysis.
Arquivo Brasileiro de Medicina Veterinária e Zootecnia,
Vol. 74,
Issue. 5,
p.
901.
Mabunda, Ripfumelo Success
Makgahlela, Mahlako Linah
Nephawe, Khathutshelo Agree
and
Mtileni, Bohani
2022.
Evaluation of Genetic Diversity in Dog Breeds Using Pedigree and Molecular Analysis: A Review.
Diversity,
Vol. 14,
Issue. 12,
p.
1054.
Bramante, Grazia
Pieragostini, Elisa
and
Ciani, Elena
2022.
Genetic Variability within the Murgese Horse Breed Inferred from Genealogical Data and Morphometric Measurements.
Diversity,
Vol. 14,
Issue. 6,
p.
422.
Ivanković, Ante
Bittante, Giovanni
Šubara, Gordan
Šuran, Edmondo
Ivkić, Zdenko
Pećina, Mateja
Konjačić, Miljenko
Kos, Ivica
Kelava Ugarković, Nikolina
and
Ramljak, Jelena
2022.
Genetic and Population Structure of Croatian Local Donkey Breeds.
Diversity,
Vol. 14,
Issue. 5,
p.
322.
de Oliveira Bussiman, Fernando
Carvalho, Rachel Santos Bueno
Ventura, Ricardo Vieira
Mattos, Elisângela Chicaroni
Ferraz, José Bento Sterman
Eler, Joanir Pereira
Fonseca e Silva, Fabyano
and
Balieiro, Júlio César de Carvalho
2022.
Founder analysis and family identification in the current Campolina horse population.
Livestock Science,
Vol. 255,
Issue. ,
p.
104796.
Rogic, Biljana
Strbac, Ljuba
Preradovic, Sladjana
and
Vazic, Bozo
2022.
Pedigree analysis of the Lipizzan horse populations from Bosnia and Herzegovina and Serbia: Structure, inbreeding and genetic variability.
Czech Journal of Animal Science,
Vol. 67,
Issue. 12,
p.
483.
Klein, Renáta
Oláh, János
Mihók, Sándor
and
Posta, János
2022.
Pedigree-Based Description of Three Traditional Hungarian Horse Breeds.
Animals,
Vol. 12,
Issue. 16,
p.
2071.
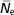 $$ \hskip3pt{\bar{\hskip-2ptN}_{{\rm e}} } $$
), and paired increase in coancestry
$$ \hskip3pt{\bar{\hskip-2ptN}_{{\rm e}} } $$
), and paired increase in coancestry 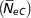 $$\left(\! \hskip3pt{\bar{\hskip-2ptN}_{{ eC}} } \right)$$
, were 74.4±3.9 and 73.5±0.58, respectively. The inbreeding increases with the pedigree knowledge. In addition, the decrease in inbreeding in last years is more noticeable.
$$\left(\! \hskip3pt{\bar{\hskip-2ptN}_{{ eC}} } \right)$$
, were 74.4±3.9 and 73.5±0.58, respectively. The inbreeding increases with the pedigree knowledge. In addition, the decrease in inbreeding in last years is more noticeable.

