Introduction
Napier grass (Pennisetum purpureum) is a perennial tropical grass and is considered one of the most important forage for ruminant production (Guan et al. Reference Guan, Shuai and Ran2020). Its highly efficient C4 photosynthetic pathway enables fast growth and high biomass yields. Similar to other tropical forage, Napier grass is cultivated in tropical and sub-tropical areas such as southern China (Dong et al. Reference Dong, Li and Wang2022a). The climate of these areas is characterized by coinciding rainy and hot seasons, enabling seasonal growth and harvest of Napier grass (Du et al. Reference Du, Yamasaki and Oya2024). Therefore, storing Napier grass biomass is an important means of maintaining the supply feed for ruminant livestock.
Ensiling can preserve green forage nutrients in a long-term and stable manner because of its anaerobic conditions, lactic acid (LA) production via water-soluble carbohydrate (WSC) fermentation by LA bacteria, and acidic environment that inhibits the growth of undesirable microorganisms (Muraro et al. Reference Muraro, De Almeida Carvalho‐Estrada and Oliveira Pasetti MH2021b). The fermentation quality of silage is influenced by environmental conditions, chemical composition, and epiphytic microorganisms of forage (Zhao et al. Reference Zhao, Jing and Yin2023). Regarding microorganisms, even if forage is harvested on the same day, the quality of fermentation can be affected by the epiphytic microorganisms of the forage (Dong et al. Reference Dong, Li and Wang2022b). Dong et al. (Reference Dong, Li and Wang2022a) reported that the time of harvest has an effect, as Napier grass harvested in the afternoon results in poor silage fermentation quality, mainly due to differences in the epiphytic microorganisms of the forage. Epiphytic microbial communities of forage are the source of microorganisms in natural silage (Zhao et al. Reference Zhao, Yin and Wang2021). Microbial communities in high-quality fermentation silage are dominated by LA bacteria, whereas silage aerobic stability is associated with fungi (molds and yeasts) (Guan et al. Reference Guan, Shuai and Ran2020). Therefore, analyzing the dynamics of microbial community structure during the process from forage to ensiling to air exposure of silage is essential to better understand the fermentation profiles of Napier grass.
However, a systematic microbial analysis of Napier grass silage remains to be reported. We hypothesize that the same species of Napier grass at the same period of growth may still exhibit different epiphytic microbial communities due to regional factors, further leading to variable fermentation quality of Napier grass silage. To test our hypothesis, Napier grass cultivated in three regions was harvested for silage during the same growth period. This study investigated the fermentation quality, aerobic stability, and microbial community structure of Napier grass silage from three regions. The results provide a reference for analyzing the microbial community structure and production of silage of high fermentation quality.
Materials and methods
Silage preparation
The Napier grass (Pennisetum purpureum Schum.) used in this study was cultivated in three areas in Nanning City, Guangxi Province, China. The sampling locations were Guangxi Xingtaimin Agricultural Company (22°68ʹN, 108°01ʹE; NGP1), the experimental farm of Guangxi University (22°83ʹN, 108°29ʹE; NGP2), and Guangxi Buffalo Breeding Stock Farm (22°89ʹN, 108°35ʹE; NGP3). Napier grass was manually harvested at the first cutting and maturity stage, leaving a stubble of approximately 10 cm on July 15, 2021. After harvest, Napier grass from various regions was immediately chopped into pieces 2–3 cm in length with a forage cutter (model zfd5570, Zheng Feng Machinery Company, China). First, the stems and leaves of chopped Napier grass were mixed thoroughly. Then, 2 kg of the chopped Napier grass was weighed using an electronic scale, after which compacted grass was placed into 2.5 L laboratory silos layer by layer using a wooden stick. The silos were stored at ambient temperature (25 ± 2.5°C) after being sealed with plastic cover and plastic tape. Four silo replicates from each location (NGP1, NGP2, and NGP3) were opened after 60 days of ensiling to assess chemical composition, microbial populations, and aerobic stability.
Sample preparation and chemical analyses
After 60 days of ensiling, the silage from each silo was emptied into a clean plastic basin and divided into three sub-samples. The first sub-sample (300 g) was oven-dried for 72 h at 65°C and then used to determine air-dry matter. The dried samples were ground and passed through a 1 mm sieve to determine the nutritional composition of Napier grass silage. The neutral detergent fiber (NDF) and acid detergent fiber (ADF) of Napier grass were determined with a fiber analyzer (ANKOM Technology, Macedon, NY, USA) according to the method described by Van Soest et al. (Reference Van Soest, Robertson and Lewis1991). Total nitrogen of Napier grass was determined with a Kjeldahl apparatus (Kjeltec 8200; FOSS, Sweden) using the Kjeldahl method, and crude protein content was calculated by multiplying total nitrogen by 6.25 (Krishnamoorthy et al. Reference Krishnamoorthy, Muscato and Sniffen1982). WSC content was analyzed with anthrone sulfuric acid colorimetry (Krishnamoorthy et al. Reference Krishnamoorthy, Muscato and Sniffen1982).
The second sub-sample (20 g) was immersed in distilled water (120 mL) at 4°C for 24 h, and then the solution was filtered through filter paper and two layers of gauze to obtain the filtrate. The filtrate was used to determine organic acids, ammonia–N (NH3–N) contents, and pH according to the specific procedure employed by Gu et al. (Reference Gu, Zhang and Zhou2023). The pH of the filtrate was measured with an electrode pH meter (HANNA pH 211, Hanna Instruments, Padova, Italy). The NH3–N content was measured with the phenol-hypochlorite reaction using the method of Broderick and Kang (Reference Broderick and Kang1980). The contents of ethanol, LA, and acetic, propionic, and butyric acid were determined by gas chromatography (Agilent 1260; Agilent Technologies, Germany) described by Zhang et al. (Reference Zhang, Zhou and Gu2019). The last sub-sample was used to determine aerobic stability and for microbiological analysis.
Aerobic stability test
The aerobic stability of Napier grass silage was tested using the same method used by Nishino and Touno (Reference Nishino and Touno2005). A Napier grass silage sample (1 kg) from each silo was loosely filled into a plastic bucket, covered with two layers of gauze, and left at room temperature (25 ± 2°C). After 1, 3, 5, and 7 days of air exposure, silage samples were thoroughly mixed and sub-sampled for analyses of pH, organic components, ethanol, and microbial communities.
Microbial community analyses
To analyze the epiphytic microbial communities of fresh, silage, and air-exposed silage samples of Napier grass, microbial DNA extraction was conducted according to the methods of Zhang et al. (Reference Zhang, Zhou and Gu2019). Microbial DNA was sent to Huada Genome Sequencing Company (Shenzhen, China) for 16S rRNA and ITS-1 gene amplicon sequencing on an Illumina MiSeq PE250 platform. The universal primers 338F (5′-ACTCCTACGGGAGGCAGCAG-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT3′) were used to amplify sequencing of the bacterial 16S rRNA V3–V4 regions. Amplification sequencing of fungal ITS was performed using the primers ITS1F (5′-CTTGGTCATTTAGAGGAAGTAA-3′) and ITS2-2043R (5′-GCTGCGTTCTTCATCGATGC-3′). Next, purified Polymerase Chain Reaction (PCR)amplicons were paired-end sequenced using the Illumina MiSeq PE300 platform (Illumina Inc., San Diego, CA, USA). Paired-end reads were merged and checked by FLASH (Version 1.2.11), and the primers were trimmed using QIIME II. Operation taxonomic units were done using open reference clustered with a 97% similarity cut-off using UPARSE (Version 7.1; http://drive5.com/uparse/), and the chimeric sequences were identified and removed using Amplicon Sequence Variant (ASV). Alpha-diversity estimates and beta-diversity evaluation, based on principal coordinate analysis (PCoA), were performed using the Phyloseq and Vegan packages in R. Bacterial and fungal communities at the phylum and genus levels were analyzed based on the Silva database with a confidence threshold of 70%. The high-throughput sequencing data were analyzed on the free online platform of Majorbio I-Sanger Cloud Platform (www.i-sanger.com).
Statistical analysis
Statistical Packages for Social Science software (IBM SPSS 20 for Windows) was employed for the statistical analyses. One-way analysis of variance was performed on each parameter of the fresh Napier grass and silage, and a general linear model was used for a two-way analysis of variance for silage air exposure days, different sampling locations, and their interaction. The different sample means were compared for significance by Duncan’s multiple range method, and significance was declared at P < 0.05. The PICRUSt2 functional prediction and co-occurrence network analyses of bacterial communities were predicted by the Cloud Platform (bioincloud.tech). The data were visualized using GraphPad Prism 9.0 software.
Results and discussion
Chemical characteristics of fresh and ensiled Napier grass
The chemical characteristics of fresh and silage Napier grass are shown in Table 1. There were significant differences (P < 0.05) in the DM, NDF, ADF, and WSC contents of fresh Napier grass samples from different sampling locations. WSCs are one of the key factors determining the quality of silage fermentation, as they can provide fermentation substrates for epiphytic microorganisms (Gomes et al. Reference Gomes, Bueno and Osmari2021). In this study, although the Napier grass from the three sampling areas was of the same cultivar and growth period, it contained significantly different carbohydrate contents. These differences can be attributed to variable soil fertility and water levels, indicating that variations in cultivated land can affect the chemical composition of forage (Kung et al. Reference Kung, Shaver and Grant2018). LA bacteria are critical beneficial microorganisms during ensiling, as they can inhibit spoilage microorganisms by fermenting WSCs to produce LA. LA has a lower pKa value compared with other organic acids such as acetic and propionic acid, which means that LA is the most effective organic acid for decreasing pH during ensiling (Yin et al. Reference Yin, Long and Zhao2023). Thus, the lowest pH of NP3 silage is related to the higher carbohydrate content of fresh Napier grass. All silage samples had lower DM weight and WSC and crude protein content compared with fresh samples, and this can be attributed to plant cell respiration during the early ensiling stages and microbial fermentation during later ensiling stages (Zhang et al. Reference Zhang, Ding and Usman2023a). After ensiling, NDF and ADF content increased, which could be explained by the loss of DM, including crude protein and WSCs. Lignocellulose is not easily degraded by microorganisms during natural ensiling, resulting in increased relative contents of neutral and ADF, as consistently reported by Zhao et al. (Reference Zhao, Dong and Li2019) and Li et al. (Reference Li, Ding and Zhao2022). Appropriate moisture and DM content are crucial for high-quality ensiling. Insufficient moisture levels inhibit the growth of LA bacteria, whereas excessive amounts of water can lead to the fermentation of spoilage microorganisms (e.g., clostridial or mycotic fermentation) resulting in loss of crude protein and WSCs (Wang et al. Reference Wang, Cai and Shao2022a). Thus, the highest pH value (5.19) of NGP1 may be due to the highest moisture levels of the fresh samples (81.27%).
Table 1. Chemical components of Napier grass from different areas before and after ensiling

a-c Means with different lowercase letters in the same row differ at P < 0.05.
Note: FW: Fresh weight; DM: dry matter; NDF: neutral detergent fiber; ADF: acid detergent fiber; CP: crude protein; WSC: water-soluble carbohydrate; LAB: lactic acid bacteria; BC: buffering capacity; Fresh: Napier grass before ensiling.
Fermentation characteristics of Napier grass silage before and after air exposure
The air exposure days, sampling location, and their interaction had significant (P < 0.05) effects on the fermentation of Napier grass silage (Table 2). After ensiling, the pH of NGP3 (3.43) was significantly lower (P < 0.05) than that of NGP2 (4.66) and NGP1 (5.19). The main reason for this is related to the high WSC content in NGP3, which provided sufficient substrate for LA production by LA bacteria and thus lowered the pH (Muck et al. Reference Muck, Nadeau and McAllister2018). During air exposure, pH mostly increased, and LA concentration decreased in all groups. This is due to LA assimilation that is used by surface aerobic spoilage epiphytic microorganisms (yeasts and molds) to propagate under aerobic conditions (Borreani et al. Reference Borreani, Tabacco and Schmidt2018). Interestingly, NGP3 showed a dramatic increase in pH (from 3.43 to 7.41) and a decrease in LA concentration (from 58.64 to 7.67 g/kg DM) compared with NGP2 (from 4.66 to 5.30 and 12.87 to 5.74 g/kg DM) and NGP1 (from 5.19 to 5.69 and 8.24 to 6.38 g/kg DM) during 7 days of air exposure. This indicates that silage quality cannot be judged unilaterally from post-silage pH and LA concentration. The concentrations of acetic and propionic acid of NGP1 were significantly higher (P < 0.05) than those of NGP2 and NGP3 after ensiling. During air exposure, the acetic and propionic acid concentrations showed an increasing tendency, and the concentrations of acetic and propionic acid of NGP1 remained consistently higher than those of NGP2 and NGP3. Acetic acid can improve the aerobic stability of silage by inhibiting the activity of aerobic spoilage microorganisms such as yeast, mold, and Clostridium (Du et al. Reference Du, Sun and Chen2021). This explains why the pH of NGP2 did not significantly decrease, possibly because acetic acid was maintained at a higher concentration during air exposure. A previous study reported that acetic acid is mainly produced by Acetobacter aceti and heterotrophic fermentation of LA bacteria in silage (Bai et al. Reference Bai, Rafiq and Li2021). The acetic acid concentration of NGP1 was significantly higher than that of NGP2 and NGP3, which may be related to variations in epiphytic microbiota and the fermentation type of LA bacteria of Napier grass silage (Du et al. Reference Du, Yamasaki and Oya2024).
Table 2. Fermentation characteristics of Napier grass silage from different regions and during air exposure
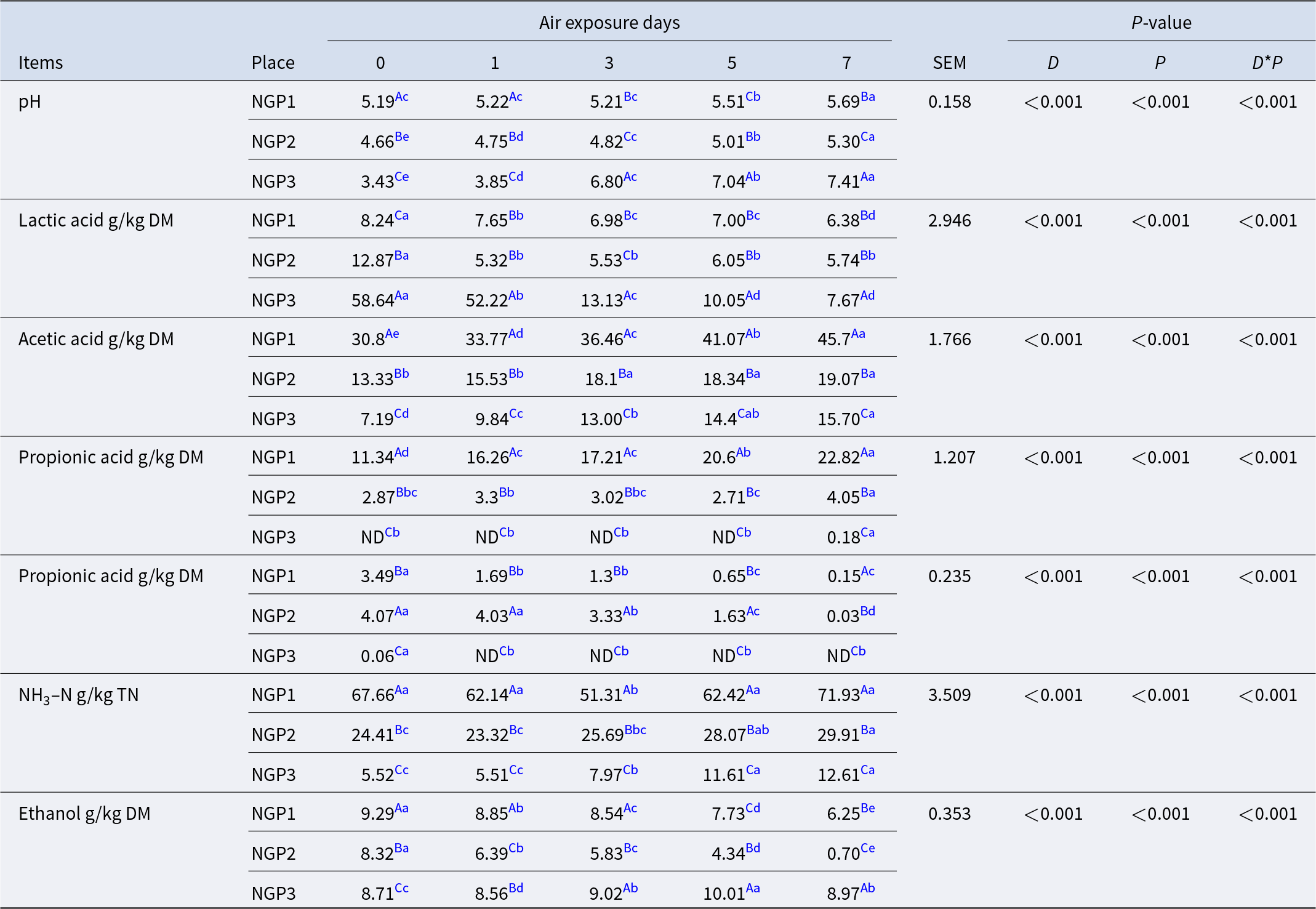
a-e Means with different lowercase letters in the same row differ at P < 0.05.
A−C Means with different uppercase letters in the same arrange differ at P < 0.05.
Note: DM: dry matter; NDF: neutral detergent fiber; ADF: acid detergent fiber; CP: crude protein; WSC: water-soluble carbohydrate; LAB: lactic acid bacteria; BC: buffering capacity; TN: total nitrogen; DM: dry matter.
Butyric acid and NH3–N are mainly produced by Clostridium fermentation of WSC and crude protein, leading to nutrient loss in silage (Li et al. Reference Li, Jiang and Zheng2020). However, Clostridium is not acid-resistant or anaerobic, and thus its activity is generally inhibited at low pH. NGP1 had significantly higher concentrations of butyric acid and ammonia nitrogen than those of NGP2 and NGP3, which may be explained by higher pH values and insufficient LA fermentation after 60 days of ensiling. The NH3–N concentration of NGP3 was significantly (P < 0.05) lower than that of NGP2 and NGP1 after silage. The NH3–N concentration gradually increased with air exposure days, and NGP1 had significantly (P < 0.05) higher NH3–N concentration than that of NGP2 and NGP3 after 7 days of air exposure. The increased NH3–N concentration in all samples indicates the reactivation of aerobic Clostridium, leading to nutrient loss during air exposure. A previous study reported that yeast is the main cause of silage spoilage during air exposure because it can ferment LA and WSC. This increases pH and temperature, which might improve the growth of undesirable microorganisms, such as Escherichia coli, Listeria, and Clostridium (Borreani et al. Reference Borreani, Ferrero and Nucera2019). During air exposure, ethanol is mainly produced by yeast in silage. The ethanol concentration of NGP1 and NGP2 was significantly lower than that of NGP3 after 7 days of air exposure, which may be due to their appropriate concentrations of acetic acid and LA. Kung et al. (Reference Kung, Shaver and Grant2018) indicated that moderate acetic acid concentration in silage is acceptable, as it inhibits yeast viability and thus improves aerobic stability. All fermentation parameters indicate that microorganisms are also a determining factor for silage fermentation quality and aerobic stability, and thus it is necessary to investigate the epiphytic microbiota of Napier grass silage.
Microbial diversity of fresh ensiled Napier grass and after air exposure
The alpha diversity of the bacterial and fungal communities in fresh ensiled Napier grass and after air exposure is illustrated in Fig. 1. All silage samples had lower alpha diversity indices compared with fresh samples. This was mainly due to the anaerobic and acidic conditions that inhibit the growth of some microorganisms after ensiling, thereby decreasing microbial community diversity (Oliveira et al. Reference Oliveira, Weinberg and Ogunade2017). The discrepancy in microbial species from fresh samples from different areas could be ascribed to climate conditions and chemical compositions of Napier grass (Muraro et al. Reference Muraro, De Almeida Carvalho‐Estrada and de Oliveira Pasetti2021a). Guan et al. (Reference Guan, Shuai and Ran2020) and Dong et al. (Reference Dong, Li and Wang2022b) reported that the epiphytic microorganisms of Napier grass harvested at different developmental stages or at different times of the day varied due to differences in forage chemical composition and climate (e.g., light, temperature, and moisture).
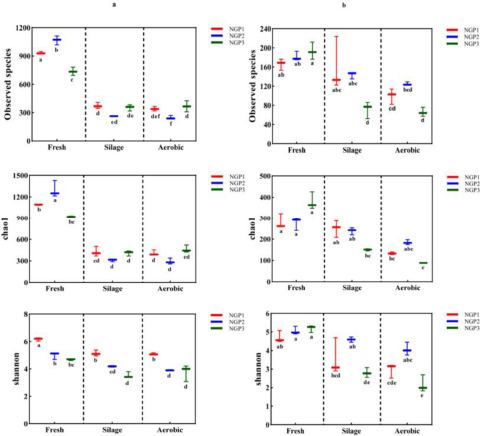
Figure 1. The alpha diversityindices of bacterial (a) and fungal (b) communities in fresh, silage, and air-exposed Napier grass samples.
The beta diversity of the microbial community in fresh ensiled Napier grass and after air exposure is illustrated by the PCoA plot (Fig. 2). A clear distinction can be observed between the fresh ensiled Napier grass and the grass after air exposure. The PCoA1 axis explained 86.51% and 76.6% of the difference in bacteria and fungi, respectively. This indicated that the variance of the original epiphytic microbial communities in forage may not change due to ensiling, which could further lead to different fermentation qualities in silage samples. This indicated that the variance of original epiphytic microbial communities in forage may not change due to ensiling, which could further lead to different fermentation qualities of silage samples.
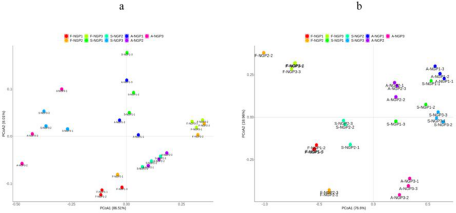
Figure 2. Principal coordinate analysis of bacteria (a) and fungi (b) from different regions after ensiling and after air exposure of Napier grass silage.
Microbial communities of fresh ensiled Napier grass and after air exposure
The microbial abundance of fresh ensiled or air-exposed samples is illustrated at the phylum and genus levels in Fig. 3. Firmicutes and Proteobacteria were predominant in all samples (Fig. 3a). Firmicutes and Proteobacteria played important roles in silage. Under anaerobic conditions, Firmicutes (e.g., Lactobacillus, Weissella, and Lactococcus) can hydrolyze WSC to produce volatile fatty acids that decrease pH; however, Proteobacteria (Pantoea, Pseudomonas, and Enterobacter) are not resistant to acids, and their growth is inhibited under anaerobic conditions (McAllister et al. Reference McAllister, Dunière and Drouin2018). In this study, the abundance of Firmicutes and Proteobacteria increased and decreased, respectively, in all samples after ensiling, indicating that Firmicutes are more competitive than Proteobacteria during ensiling. The decrease in Proteobacteria abundance favors the improvement of silage nutrients and fermentation quality because Proteobacteria can accelerate carbon and nitrogen cycling during silage, decreasing DM (Borreani et al. Reference Borreani, Tabacco and Schmidt2018). In addition, the lower abundances of Pantoea, Pseudomonas, and Enterobacter in silage could prevent intestinal diseases in animals.
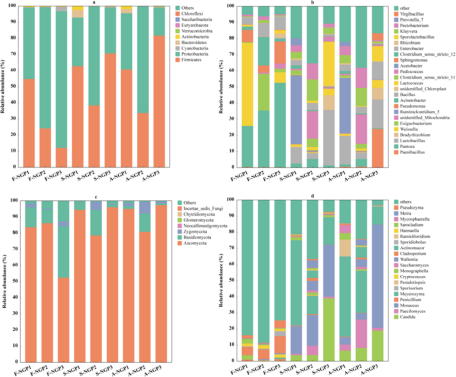
Figure 3. The relative abundances of bacterial and fungal communities of Napier grass from different regions after ensiling and after air exposure at the phylum level (a, c) and the genus level (b, d).
Lactococcus and Lactobacillus can use one molecule of glucose to produce two molecules of LA, which can rapidly acidify and decrease the pH of silage (Guo et al. Reference Guo, Xu and Li2023). In this study, the highest LA concentration and lowest pH in SNGP3 may be attributed to the relatively high abundance of Lactobacillus and Lactococcus. High moisture levels (>80%) and lack of WSC can lead to inadequate fermentation by LA bacteria, preventing the decrease in pH to a suitable range and resulting in poor-quality silage fermentation (Zhang et al. Reference Zhang, Ding and Usman2023b). However, Clostridium can develop a competitive advantage under such conditions (Xu et al. Reference Xu, Li and Wang2023a), which explains the predominant abundance of Ruminiclostridium_5 (42.35%) detected in S-NGP1. The prolonged presence of Clostridium during ensiling could accelerate WSC and crude protein degradation, decreasing WSC content and increasing NH3–N concentrations, as detected in NGP1. Yeasts and molds are considered the main microorganisms responsible for the deterioration of grass silages upon exposure to air because they can withstand organic acids better than most other microorganisms under aerobic conditions (Gallo et al. Reference Gallo, Minuti and Bani2020). Moreover, they can oxidize organic acids and other chemical compositions of silage to produce ethanol and mycotoxins, thus contaminating silage (Bernardes et al. Reference Bernardes, Daniel and Adesogan2018). A-NGP3 had higher abundances of Candida (18.97%) and Monascus (63.98%) compared with NGP1 (6.58% and 4.57%) and NGP2 (8.11% and 4.03%). This may have caused the sharp increase in pH and rapid decrease in LA concentration of A-NGP3 during air exposure. This indicates that high LA concentration and low pH are not the only indicators of silage fermentation quality. The same conclusion was reported by Wang et al. (Reference Wang, Dong and Li2019). Interestingly, Meyerozyma abundance predominated (>10%) in all silage and air-exposed samples but was lower (<1.06%) in the fresh samples. Few reports have discussed this phenomenon. Keshri et al. (Reference Keshri, Chen and Pinto2018)found a higher abundance (53.5%) of Meyerozyma in fresh samples that decreased after fermentation. Further research is needed to determine the effect of Meyerozyma on fermentation quality and the aerobic stabilization of forage silage.
Correlation analysis of fermentation parameters and microbial communities
The correlations between fermentation parameters and microbial communities of silage or air-exposed samples were analyzed using Spearman’s correlation heatmaps (Fig. 4). The pH and NH3–N concentration was negatively correlated with Lactococcus (S-NGP3, P < 0.05). Lactococcus plays a crucial role in tropical forage silage (Oliveira et al. Reference Oliveira, Weinberg and Ogunade2017). Lactococcus are homofermentative LA bacteria that can ferment 1 mol of glucose to produce 2 mol of LA (Okoye et al. Reference Okoye, Wang and Gao2023), which means that they ferment glucose more efficiently than heterofermentative LA bacteria. In S-NGP3, Lactococcus was positively correlated with LA concentration and negatively correlated with pH, which may underly the rapid production of LA. This was consistent with the lowest pH and highest LA concentration in S-NGP3 after ensiling. In A-NGP2, the positive correlation between ethanol and Meyerozyma may be due to the fact that Meyerozyma has anaerobic tolerance and can ferment WSC and VFA to produce ethanol in silage. Wang et al. (Reference Wang, Zhang and Zhao2023) reported that Sphingomonas causes hydrolysis of soluble protein in agricultural by-products and red clover silage. NH3–N was mainly produced by protein hydrolysis during silage (Benjamim Da Silva et al. Reference Benjamim Da Silva, Polukis and Smith2024), thus the positive correlation between Sphingomonas and NH3–N in all silage samples. Acetic acid can inhibit fungal growth during air exposure to improve the aerobic stability of silage (Wang et al. Reference Wang, Zhang and Zhao2023). Acetic acid was negatively correlated with Candida and Monascus in A-NGP3, which indicates that these species may play a pivotal role in promoting aerobic corruption of silage. Wang et al. (Reference Wang, Nie and Tian2022b) also found higher abundances of Candida and Monascus in whole-crop maize during air exposure.

Figure 4. Correlation heatmaps of fermentation parameters and epiphytic bacterial (a, b) and fungal (c, d) microbiota (top 23 genera) in Napier grass silage and after air exposure. Red squares represent positive correlations (0 < r < 1), and blue squares represent negative correlations (−1 < r < 0). *: significant at P < 0.05; **: significant at P < 0.01.
Metabolic prediction and co-occurrence networks of bacterial communities
KEGG and co-occurrence networks can further illustrate microbial community metabolic pathways and interactions (Wang et al. Reference Wang, Nie and Tian2022b). The relative abundances of epiphytic bacterial community KEGG level-1 and level-2 pathways were predicted by PICRUSt2 (Fig. 5). Metabolism was the dominant level-1 metabolic pathway (>70%) in all fresh, silage, and air-exposed samples. Carbohydrate metabolism, amino acid metabolism, metabolism of cofactors and vitamins, and metabolism of other amino acids were the main level-2 pathways, all of which belong to level-1 of metabolism with a relative abundance of more than 6%. Carbohydrate metabolism was the most abundant metabolic pathway and increased after ensiling and air exposure (11.01–11.55%), compared with fresh samples (10.35–10.38%). Carbohydrate metabolism was the predominant metabolic pathway, suggesting that carbohydrate metabolism is the primary pathway for sustaining metabolism (Xu et al. Reference Xu, Li and Wang2023a). Carbon metabolism pathways were increased after silage, probably due to the predominance of WSCs fermentation by LA bacteria (Du et al. Reference Du, Yamasaki and Oya2024). Protein hydrolysis can increase amino acid metabolism and decrease the abundance of the main microorganisms involved in the nitrogen cycle after silage (Proteobacteria Pseudomonas and Enterobacter). These species may decrease amino acid metabolic pathways (Bai et al. Reference Bai, Ding and Su2022). In all silage and air-exposed samples, amino acid metabolism and chemical structure transformation maps of metabolic pathways were decreased compared with fresh samples.

Figure 5. Changes of KEGG metabolic pathways on levels 1 and 2 obtained with PICRUSt2 from fresh, silage, and air-exposed Napier grass samples. KEGG, Kyoto Encyclopedia of Genes and Genomes.
The networks of bacterial communities in fresh, silage, and air-exposed samples are shown in Fig. 6. The raw materials from different areas exhibited distinct co-occurrence patterns. Compared to fresh samples, silage and air-exposed samples exhibited more complex co-occurrence networks of bacterial communities. The silage sample S-NGP1 had more nodes and edges in bacterial networks (155) than S-NGP2 (96) and S-NGP3 (79). Additionally, the air-exposed sample A-NGP3 had a higher proportion of positive correlations (94.79%) than either A-NGP1 (58.62%) or A-NGP2 (47.42%). Zhao et al. (Reference Zhao, Yin and Li2022) analyzed the co-occurrence networks of the ten most abundant epiphytic microorganisms on the surface of sweet sorghum and found that the co-occurrence networks became simpler after silage. However, this study found that the co-occurrence networks of Napier grass epiphytic microorganisms became complex, probably because the top 20 microorganisms were selected for analysis in this study. Despite the decrease in microbial diversity after ensiling, the co-occurrence networks in S-NGP1 were more complex than those in S-NGP2 and S-NGP3. This may be due to the higher pH, which resulted in more active bacteria in S-NGP1. In A-NGP3, bacteria were found to have an extremely high ratio of positive co-occurrence network relationships (94.79%) compared with A-NGP2 (47.42%) and A-NGP1 (58.62%); this phenomenon has not been reported in previous studies. We hypothesize that the higher fermentation quality of NGP3 may be related to the fact that fermentation products, such as LA and WSCs, provide sufficient nutrients for bacteria, making them less competitive and more cooperative (Penagos-Tabares et al. Reference Penagos-Tabares, Mahmood and Khan2023). Future studies are needed to further analyze this phenomenon.

Figure 6. Co-occurrence network analyses of fresh, silage, and air-exposed samples of Napier grass from different regions. (a) Fresh samples; (b) silage samples; and (c) air-exposed samples. Nodes represent bacterial genera, and node sizes represent their relative abundance. The colors of the connecting lines between nodes represent positive correlations (red) and negative correlations (blue) of bacterial genera.
Conclusion
This study analyzed the fermentation patterns, bacterial communities, and aerobic stability of Napier grass silage from various regions. The fermentation quality and aerobic stability of Napier grass silage from different regions are driven by epiphytic microbiota. A high relative abundance of Lactococcus in the epiphytic microbiota of fresh Napier grass improves the fermentation quality of silage. In contrast, higher relative abundances of Candida and Monascus in Napier grass silage accelerate aerobic spoilage.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.
Author contributions
HD: methodology, formal analysis, visualization, writing-original draft preparation. HD and QY: validation, methodology, software. NZ: investigation, formal analysis. QG and QT: methodology, visualization. CZ and BL: conceptualization, project administration, resources, data curation, supervision, funding acquisition, writing—review and editing. All authors read and approved the final manuscript.
Funding statement
This work was funded by the National Natural Science Foundation of China (Grant No.312360851) and National Key Research and Development Program of China [grant number 2022YFD1300602].
Conflict(s) of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.










