Introduction
Lactation persistency is an important economic indicator in dairy cows, referring to the rate of milk yield decline after the peak of lactation, which determines the annual yield and mammary milk yield maintenance ability of dairy cows (Cesarani et al. Reference Cesarani, Gaspa, Masuda, Degano, Vicario, Lourenco and Macciotta2020; Dekkers et al. Reference Dekkers, Ten Hag and Weersink1998). The decrease of lactation persistency is related to poor mammary gland health status of dairy cows, such as mammary diseases and metabolic stress (Harder et al. Reference Harder, Bennewitz, Hinrichs and Kalm2006), and low feed efficiency (Solkner and Fuchs Reference Solkner and Fuchs1987). Therefore, a higher lactation persistency can prevent a rapid milk loss after peak point. The number/density of mammary epithelial cells and cell viability are essential to sustain a high lactation persistency, which is determined by a balance of cell proliferation and apoptosis. When proliferation and apoptosis are unbalanced, the lactation persistency would be deteriorated (Boutinaud et al. Reference Boutinaud, Herve and Lollivier2015; Stefanon et al. Reference Stefanon, Colitti, Gabai, Knight and Wilde2002). Studies clearly stated that reduced mammary cells caused by increased apoptosis rate play predominant role in lactation persistency (Knight and Peaker Reference Knight and Peaker1984; Yart et al. Reference Yart, Dessauge, Finot, Barbey, Marnet and Lollivier2012).
Factors including nutrients, environmental change and redox status play crucial roles in modulating cell apoptosis/proliferation and thus affect lactating persistency (Stefanon et al. Reference Stefanon, Colitti, Gabai, Knight and Wilde2002). Previous studies mainly focused on the genetic aspects of the prediction of lactation persistency (Cole and Null Reference Cole and Null2009), whereas limited information is available in physiological perspective. As an important physiological factor, oxygen plays a central role in intracellular adenosine triphosphate (ATP) production and acts as a major electron acceptor in various biochemical reactions (Semenza Reference Semenza2010). Hypoxia-inducible factor 1α (HIF-1α) is an important transcription factor that senses hypoxia and involved in multiple biological processes, such as tissue development, environmental adaptation, redox alteration and tumor inhibition (Kaelin and Ratcliffe Reference Kaelin and Ratcliffe2008; Lee et al. Reference Lee, Chandel and Simon2020). Other studies suggest that activating HIF-1α is involved in accumulating reactive oxygen species (ROS) in the tissue and thus potentially leads to health risk of tissues with high metabolic activity (Moley and Mueckler Reference Moley and Mueckler2000; Pialoux and Mounier Reference Pialoux and Mounier2012). For example, elevated HIF-1α accumulation in the mammary gland during lactation stage due to higher oxygen consumption was responsible for changes in energy metabolism (Seagroves et al. Reference Seagroves, Hadsell, McManaman, Palmer, Liao, McNulty, Welm, Wagner, Neville and Johnson2003). As a tissue with high metabolic activity, the relationship between oxygen metabolism and lactation capacity of the mammary gland in dairy cows is not clear yet.
We hypothesized that cows with different lactation persistency are associated with their oxygen metabolism status in the mammary gland. To test this hypothesis, a dairy cow trial was designed, in which a group of cows were selected and sampled at different time points from the peak lactation to explore the association between hypoxia and lactation persistency and the underlying mechanism. Our study provides a new perspective to explore the issue of lactation persistency and will provide a theoretical basis and potential strategies to help improve annual yield and maintain mammary gland health in dairy cows.
Materials and methods
Animal trials
Animals and management
All experimental procedures were approved by the Animal Use and Care Committee of Zhejiang University (Hangzhou, China). At the beginning of the trial, a total of 75 healthy, multiparous, high-yielding Holstein dairy cows with similar days in milk (DIM) and milk yield at peak lactation were selected. The total trial period lasted 180 days, covering the peak, middle and late lactation to obtain lactation persistency of dairy cows. Their health was monitored by veterinarians on the farm throughout the trial. Specifically, veterinarians conducted daily disease examination and diagnosis, and cows that develop diseases were given symptomatic treatment or eliminated. After monitoring the health of 75 cattle for 180 days, 14 of them were eliminated because they developed health problems, including mastitis, hoof disease, digestive disease, etc. Ultimately, 61 cows remained healthy throughout the trial and were used in the current study. The 61 cows remained similar in terms of parity (2.5 ± 0.8), DIM (88 ± 4.3) and milk yield (44.9 ± 5.35 kg/day) at peak lactation. In order to obtain lactation persistency and lactation curve during the 180 days, milk yield of each cow was recorded every 15 days using the ApolloMilkSystem (GEA Farm Technologies, Naperville, USA).
The milk yield at each time point was derived from the average of 2 consecutive days, and then the decline curve during the 180-day milk yield from the peak lactation was obtained at a total of 13 recording points at 15-day intervals. In addition, samples of milk, blood and mammary tissues were collected at d 0, 90 and 180 (corresponding to d 88, 178 and 268 of DIM, respectively; see below). The diagram of the animal trial design was shown in Fig. 1A.

Figure 1. Experimental layout. A Diagram of in vivo studies in dairy cows. B Diagram of in vitro studies in bovine mammary epithelial cells (MAC-T).
Throughout the trial period, the cows were housed in free-stall pens and milked three times daily using an automatic milking system, including morning, noon and evening milking at 0500, 1300 and 2000 h. All animals had free access to drinking water and feed. These cows were given total mixed rations (TMRs) at 0500, 1300 and 2000 h a day to ensure that they have enough feed immediately after milking. The remaining feed was removed before the next feeding. The feed formulation was adjusted by the cattle farm according to the lactation period, which was different during the peak, middle and late lactation. Feed formulation was obtained at d 0, 90 and 180, respectively, as three key time points. The details of diet ingredients and composition are presented in Table S1. To obtain dry matter intake (DMI), each of the 61 cows was kept in an individual tie-stall for about a week ahead of d 0, 90 and 180. Feed intake was measured individually by weighing offered feed and residual feed from 2 consecutive days at d 0, 90 and 180. The corresponding DMI of individual animals was calculated by multiplying the daily feed intake with the dry matter content of the TMR.
Sample collections
Milk samples
Milk samples were collected into the centrifuge tubes using milk-sampling devices (Waikato Milking Systems NZ Ltd, Waikato, Hamilton, New Zealand) at three milking times in the morning, noon and evening. The three milk samples were added into a 50 mL centrifuge tube with a bronopol tablet (milk preservative, D & F Control Systems, San Ramon, CA, USA) in a ratio of 4:3:3 and mixed well. The samples were stored at 4°C, and the milk composition analysis was completed within 2 days.
Blood samples
Blood samples were collected from the mammary vein and caudal artery for each of the 61 cows at 3 h (0800–0930 h) after the morning feeding at d 0, 90 and 180. Blood samples from mammary vein were collected into 10 mL lithium heparin-containing vacuum tubes (10 mL, Becton Dickinson, Franklin Lakes, NJ) through individually packed blood collection needles. After the required amount of blood sample was collected to the lithium heparin vacuum tube, the needle connected to the tube was first pulled out, and then the needle inserted into the mammary vein was pulled out to avoid air entering the tubes. Two hundred microliters of blood sample in the lithium heparin vacuum tube was taken through the pipette gun and added to the blood gas card to detect the blood gas parameters immediately. The remaining sample was centrifuged at 3000 × g at 4°C for 15 min to obtain the plasma and stored at −20°C until analysis. Blood samples from the caudal artery were collected into 10 mL lithium heparin vacuum tubes using the same blood collection method as mammary vein, centrifuged under the same centrifugal conditions, and plasma was obtained and stored at −20°C until analysis.
Mammary gland tissue samples
Mammary gland tissues were obtained for each of the 61 cows by surgical procedures at d 0, 90 and 180. The surgical sites at these three time points were selected from the middle of right-rear udder, left-rear udder and right-rear udder of cows, respectively. The specific operation method of biopsy was referred to the procedures from Farr et al. (Reference Farr, Stelwagen, Cate, Molenaar, McFadden and Davis1996) with slight modifications. Briefly, the biopsy was performed about 3 h after milking in the morning. Cows were confined to separate stalls of the cattle shed. Cows were anesthetized by intramuscular injection of Sedazine II (xylazine hydrochloride injection). An area of about 10 cm2 in the right rear region of the mammary gland was selected to avoid major blood vessels. The skin surface of the area was shaved using a disposable razor (Gillette Blue II™, Gillette, Reading, UK). The skin was scrubbed and disinfected with iodine and 75% ethanol solution. Local anesthesia was performed by subcutaneous injection of procaine hydrochloride at the biopsy site. The skin and gland capsule were cut 1–2 cm with a scalpel. Then, a core (70 × 4 mm in diameter) of mammary tissue was cut with the biopsy instrument, which was rotated by a slow speed electric motor. A retractable blade of the biopsy instrument was extended to cut the inner end of the core, after which the tissue sample was withdrawn with the instrument. The collected tissues were immediately washed and trimmed in sterile physiological saline and divided into two parts. One sample was fixed in 4% paraformaldehyde and stored at 4°C for the terminal deoxynucleotidyl transferase-mediated deoxyuridine triphosphate nick end labeling (TUNEL) assays. The other sample was immediately placed in a cryopreservation tube (Corning Incorporated, Corning, NY, USA), quenched with liquid nitrogen and then stored at −80°C until analysis. After obtaining the sample, the biopsy incision of mammary gland was closed with absorbable sutures. Penicillin and streptomycin were injected intramuscularly. The cows were milked by hand every day for the next week until all the blood clots in the udder were removed.
Grouping of lactation persistency
After the 180-day trial was completed, the recorded milk yields were used to calculate the lactation persistency of each of the 61 cows according to the below calculation formula. Lactation persistency is an indicator that reflects the degree of milk yield change between two time points. The formula for calculating lactation persistency commonly used by cattle farms in Dairy Herd Improvement reports was chosen as follows: 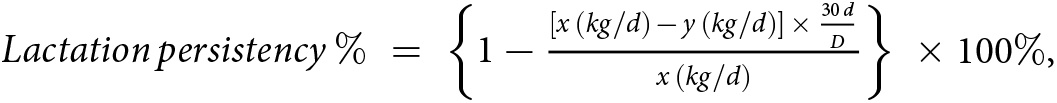 $Lactation\,persistency\,\% = \left\{ {1 - \frac{{\left[ {x\,\left( {kg/d} \right)\, - \,y\,\left( {kg/d} \right)} \right]\, \times \,\frac{{30\,d}}{D}}}{{x\,\left( {kg/d} \right)}}} \right\}\, \times 100\% $, where x and y are the milk yields corresponding to two time points (d 0 and d 180, respectively, in this study), and D is the number of days between the two time points (180 days in this experiment). The group size was determined by power analysis, which was conducted in SAS 9.4 (SAS Institute, Cary, North Carolina, USA) using α = 0.05 and one-tailed t-test. With the power level of 0.95, a group size of n = 12 was required based on lactation persistency. That is, from 61 cows, we selected top 12 cows with highest lactation persistency (95.9 ± 0.74%) and top 12 cows with lowest lactation persistency (92.2 ± 0.79%) and defined them as high lactation persistency (HP) and low lactation persistency (LP) groups, respectively.
$Lactation\,persistency\,\% = \left\{ {1 - \frac{{\left[ {x\,\left( {kg/d} \right)\, - \,y\,\left( {kg/d} \right)} \right]\, \times \,\frac{{30\,d}}{D}}}{{x\,\left( {kg/d} \right)}}} \right\}\, \times 100\% $, where x and y are the milk yields corresponding to two time points (d 0 and d 180, respectively, in this study), and D is the number of days between the two time points (180 days in this experiment). The group size was determined by power analysis, which was conducted in SAS 9.4 (SAS Institute, Cary, North Carolina, USA) using α = 0.05 and one-tailed t-test. With the power level of 0.95, a group size of n = 12 was required based on lactation persistency. That is, from 61 cows, we selected top 12 cows with highest lactation persistency (95.9 ± 0.74%) and top 12 cows with lowest lactation persistency (92.2 ± 0.79%) and defined them as high lactation persistency (HP) and low lactation persistency (LP) groups, respectively.
Sample measurements
Milk composition analysis
Milk samples were analyzed for milk compositions (protein, fat, lactose and milk urea nitrogen [MUN]) using a spectrophotometer (Foss-4000; Foss Electric A/S, Hillerod, Denmark).
Blood gas parameters
Blood gas in the mammary vein was measured via an iSTAT Portable Clinical Analyzer (Heska Corporation, Fort Collins, CO, USA). Two hundred microliters of blood samples from lithium heparin tubes was added to CG8 + cartridges (Abbott Medical, Canada) with pipette gun. Oxygen partial pressure (pO2), oxygen saturation (sO2) and hemoglobin (HGB) were obtained, and oxygen concentration (cO2) was calculated from these parameters using the formula: ![]() ${\text{c}}{{\text{O}}_2}\left( \% \right) = 0.003 \times {\text{p}}{{\text{O}}_2}\left( {{\text{mmHg}}} \right) + 1.39 \times {\text{HGB}}\left( {{\text{g}}/{\text{dL}}} \right) \times {\text{s}}{{\text{O}}_2}\left( {\text{\% }} \right){ }$ (Regli et al. Reference Regli, Hockings, Musk, Roberts, Noffsinger, Singh and Heerden2010).
${\text{c}}{{\text{O}}_2}\left( \% \right) = 0.003 \times {\text{p}}{{\text{O}}_2}\left( {{\text{mmHg}}} \right) + 1.39 \times {\text{HGB}}\left( {{\text{g}}/{\text{dL}}} \right) \times {\text{s}}{{\text{O}}_2}\left( {\text{\% }} \right){ }$ (Regli et al. Reference Regli, Hockings, Musk, Roberts, Noffsinger, Singh and Heerden2010).
Plasma oxidative stress parameters
Plasma levels of HIF-1α (#H307-2), malondialdehyde (MDA; #A003-1-2), superoxide dismutase (SOD; #A001-1-2), glutathione peroxidase (GSH-Px; #A005-1-2), total antioxidant capacity (T-AOC; #A015-3-1), heme oxygenase-1 (HO-1; #H246-1), nitric oxide (NO; #A012-1-2), endothelial nitric oxide synthase (eNOS; #H195) and inducible nitric oxide synthase (iNOS; #H372-1) were analyzed using commercial assay kits (Nanjing Jiancheng Bioengineering Institute, Nanjing, China).
Plasma biochemical parameters
Plasma concentrations of glucose, triglyceride (TG), non-esterified fatty acid (NEFA) and β-hydroxybutyric acid (BHBA) were measured using an Auto Analyzer 7020 instrument (Hitachi High-Technologies Corporation, Tokyo, Japan) with colorimetric commercial kits (Ningbo Medical System Biotechnology Co., Ltd., Ningbo, China).
Mammary tissue apoptosis assays
The TUNEL assay was used to detect mammary apoptosis. The mammary tissues were removed from 4% paraformaldehyde fixation and then dehydrated and embedded in paraffin. The paraffin-embedded tissues were sectioned to a thickness of 6 μm. After dewaxing the paraffin sections, proteinase K was added and incubated at 37°C for 20 min. Sections were washed with phosphate-buffered saline (PBS) three times for 5 min each time. The freshly prepared TUNEL reaction mixtures from the TUNEL kit (Biyuntian, Shanghai, China; # C1088) were applied to sections and incubated in a wet box at 37°C for 60 min. After washing with PBS, Hochest was added and incubated in darkness for 10 min. The sections were then washed again with PBS and sealed with anti-fluorescence quenching sealing tablets. Finally, the slices were observed under fluorescence microscopy (OLYMPUS, IX71). Hochest colored the nucleus blue, and the nuclei of TUNEL-positive cells were stained green.
Mammary gland tissue RNA sequencing
From the 2 groups of 12 cows in each group, 7 cows in each group with similar milk yield were selected for RNA sequencing, and the selection method was shown in Fig. S1. Total RNA extraction was performed using the Trizol method (Invitrogen) according to the manufacturer’s instructions. RNA degradation and contamination were examined using 1% agarose gels. RNA samples were detected for purity using the NanoPhotometer spectrophotometer (IMPLEN, CA, USA) and concentration using the Qubit RNA Assay Kit in Qubit 2.0 Fluorometer (Life Technologies, CA, USA). RNA integrity was measured using the RNA Nano 6000 Assay Kit of the Bioanalyzer 2100 system (Agilent Technologies, CA, USA). About 3 µg of total RNA per sample was used as the starting material for RNA sample preparations. Fragmentation was carried out using divalent cations under elevated temperature in NEBNext First Strand Synthesis Reaction Buffer (5X). Library fragments were purified using the AMPure XP system to preferentially obtain complementary DNA (cDNA) fragments of 250–300 bp in length (Beckman Coulter, Beverly, USA). The polymerase chain reaction (PCR) products were then purified utilizing the AMPure XP system, and the quality of the library was evaluated using an Agilent Bioanalyzer 2100 system. The index-coded samples were clustered on a cBot Cluster Generation System using the TruSeq PE Cluster Kit v3-cBot-HS (Illumina) according to the manufacturer’s instructions. After cluster generation, cDNA sequencing libraries were run on an Illumina HiSeq platform to generate 125 bp/150 bp paired-end reads.
In vitro cell assays
Cell culture and hypoxia model
The bovine mammary epithelial cell lines (MAC-T) were cultured in high-glucose Dulbecco’s modified eagle medium (HyClone; #SH30243.01) supplemented with 10% fetal bovine serum (Lonsera; #A511-001) and 1% penicillin and streptomycin (Biosharp; #BL505A). The cell concentration was adjusted to 5 × 105 cells/mL and cultured in a 6 cm cell culture dish at 37°C, 5% CO2 incubator. The cells were divided into two groups with three replicates in each group. They were placed in incubators with a low oxygen concentration of 1% and a physiological oxygen concentration of 11% (simulating venous blood oxygen concentration of dairy cows) to construct a hypoxic model. After treatment with different oxygen concentrations for 24 h, cell morphology was observed by microscope, and cell samples were collected for subsequent analyses, including apoptosis and oxidative stress. The diagram of the cell trial design was shown in Fig. 1B.
Sample measurements
Cell apoptosis assays
Cell apoptosis was measured with Hoechst Cell Apoptosis Staining Kit (Beyotime Biotechnology Company; # C0003). After the cells were rinsed with PBS, they were fixed with 4% paraformaldehyde for 15 min. The cells were washed with PBS for three times, 5 min each time, and Hoechst 33258 staining solution was applied and incubated at room temperature for 15 min away from light. After washing, images were obtained under an inverted fluorescence microscope (Olympus IX73, Olympus Corporation, Tokyo, Japan), in which apoptotic nuclei were observed by blue fluorescence.
Cellular oxidative stress indicators
The detection methods of MDA, SOD, GSH-Px and T-AOC were the same as those in the animal trial. In addition, total nitric oxide synthase (T-NOS; # A014-2-1) was determined by the commercial assay kits of Nanjing Jiancheng Bioengineering Institute. The intracellular ROS detection assay was as follows: The dichloro-dihydro-fluorescein diacetate as the probe was diluted with serum-free culture solution at 1:1000 ratio and mixed with the cells with the cell concentration of about 1–20 million per milliliter. The cell solutions were incubated for 20 min at 37°C. The cells were then washed three times with PBS and resuspended for detection of ROS by flow cytometry.
Data analysis and statistical analysis
RNA-seq data analysis
Raw data (raw reads) in the fastq format were processed using in-house Perl scripts to obtain clean data (clean reads) after removing reads containing the adapter and ploy-N and low-quality reads. Meanwhile, Q20, Q30 and the ratio of guanine to cytosine (GC) contents of the clean data were calculated. Reference genome and gene model annotation files were directly downloaded from the genome website. The reference genome index was built using Hisat2 v2.0.5, and paired-end clean reads were also mapped to the reference genome using Hisat2 v2.0.5. Read numbers were mapped to each gene and gene counts were generated using FeatureCounts v1.5.0-p3. The principal component analysis was conducted using R packages FactoMineR and factoextra. Differential gene expression was performed using edgeR. A volcano plot of differentially expressed genes (DEGs) was created using ggthemes and ggpubr R packages using P < 0.05 and |log2 (fold change)| > 1 as the screening criteria for DEGs. The Bioconductor package org.Bt.eg.db was applied to convert the bovine official gene symbols into bovine Ensembl gene IDs. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways enrichment analyses were performed using the Bioconductor R package cluster profiler. The key enriched KEGG pathways were visualized using the bubble plot created in the R package “ggplot2”. The heat map was generated with TBtools software. The correlation coefficients of two genes were analyzed using the cor function from the corrplot package. Cytoscape (version 3.2.0) was applied to visualize the gene dependency network, and topological network parameters were calculated using the Network Analyzer plug-in for Cytoscape. The network graph was visualized using Gephi software.
Statistical analysis
For animal trials, high and low lactation persistency cows were compared. Significant differences between means of lactation persistency for two groups were detected using the t-test (version 9.4; SAS Institute, Inc., Cary, NC, USA). The data of lactation performance, mammary gland apoptosis and plasma indicators were analyzed using the PROC MIXED procedure of SAS 9.4 software, including experimental day (D), lactation persistency (P) and their interaction (D × P) as the fixed effects. For in vitro trial, the differences between the 1% and 11% groups were analyzed. The difference of apoptosis and oxidative stress indexes between the two groups was analyzed by independent samples t-test (version 9.4; SAS Institute, Inc., Cary, NC, USA). The data were expressed as the mean ± standard error of the mean (SEM) and were analyzed using SAS. A P-value of less than 0.05 was considered statistically significant and P < 0.01 was very significant difference.
Results
Lactation performance
The lactation persistency of the two groups is presented in Fig. S2. The decline curve of milk yield is depicted in Fig. 2. The lactation performance of cows in HP and LP groups is shown in Table 1. No differences in DMI were found between the two groups throughout the entire experimental period (P persistency = 0.93). No differences existed in milk yield between two groups until d 105 (P d 105 = 0.03), and the difference became very significant after d 105 until the end of experiment (P < 0.01). The yields of milk fat (P d 180 < 0.01), milk protein (P d 180 < 0.01), fat-corrected milk (P d 180 < 0.01) and energy-corrected milk (P d 180 < 0.01) were lower in the LP group than in the HP group on d 180. No differences were found in the contents of fat (P persistency = 0.51), protein (P persistency = 0.41) and lactose (P persistency = 0.62) between the two groups throughout the experiment.

Figure 2. Declining milk yield of dairy cows with high (HP) and low (LP) lactation persistency over the experiment period of 180 days. n = 12. Error bars indicate standard error of the mean. *P < 0.05, **P < 0.01.
Table 1. Lactation performance of dairy cows with high and low lactation persistency at d 0, 90 and 180 of the experiment1

Notes:
1 The data are expressed as the mean ± SEM, n = 12.
a,b Means without a common superscript differ significantly between the two groups at the same time point (P < 0.05).
2 DMI, dry matter intake; FCM, fat-corrected milk, FCM = milk yield × 0.4324 + milk fat yield × 16.216; ECM, energy-corrected milk, ECM = 0.3246 × milk yield + 13.86 × milk fat yield + 7.04 × milk protein yield.
3 HP, high lactation persistency; LP, low lactation persistency; d 0, day 0 of the experiment; d 90, day 90 of the experiment; d 180, day 180 of the experiment.
4 D, day; P, persistency; D × P, interaction between day and persistency.
Plasma variables
The gas profiles in the mammary vein of cows with different lactation persistency are presented in Table 2. pO2 was lower (P persistency = 0.02) in the LP group throughout the entire experiment period and tended to be lower at d 180 (P d 180 = 0.08) in the LP group than in the HP group. The cO2 was not different between the two groups (P persistency = 0.62). In addition, the overall plasma concentration of HIF-1α across the lactation stages was higher in cows of LP group than in the HP group (P persistency < 0.01), with a significant difference observed on d 0 (P d 0 < 0.01).
Table 2. Hypoxia-related indicators of dairy cows with high and low lactation persistency at d 0, 90 and 180 of the experiment1

Notes:
1 The data are expressed as the mean ± SEM, n = 12.
a,b Means without a common superscript differ significantly between the two groups at the same time point (P < 0.05).
2 pO2, oxygen partial pressure; cO2, oxygen content; HIF-1α, hypoxia-inducible factor 1α.
3 HP, high lactation persistency; LP, low lactation persistency; d 0, day 0 of the experiment; d 90, day 90 of the experiment; d 180, day 180 of the experiment.
4 D, day; P, persistency; D × P, interaction between day and persistency.
Oxidative stress variables in the plasma of HP and LP cows are shown in Table 3. MDA levels were higher (P d 180 = 0.01) and iNOS levels were lower (P d 180 = 0.01) on d 180 in the LP group than in the HP group. In addition, the plasma GSH-Px concentration was higher in the LP group than in the HP group throughout the entire experimental period (P persistency = 0.05). No significant differences in plasma concentrations of SOD, T-AOC, HO-1, NO and eNOS were found between LP and HP groups (P persistency > 0.05).
Table 3. Plasma oxidative stress indicators of dairy cows with high and low lactation persistency at d 0, 90 and 180 of the experiment1

Notes:
1 The data are expressed as the mean ± SEM, n = 12.
a,b Means without a common superscript differ significantly between the two groups at the same time point (P < 0.05).
2 MDA, malondialdehyde; SOD, superoxide dismutase; GSH-Px, glutathione peroxidase; T-AOC, the total antioxidant capacity; HO-1, heme oxygenase-1; NO, nitric oxide; iNOS, inducible nitric oxide synthase; eNOS, endothelial nitric oxide synthase.
3 HP, high lactation persistency; LP, low lactation persistency; d 0, day 0 of the experiment; d 90, day 90 of the experiment; d 180, day 180 of the experiment.
4 D, day; P, persistency; D × P, interaction between day and persistency.
Plasma physiological and biochemical variables are shown in Table 4. Plasma concentrations of glucose (caudal artery on d 90, P persistency < 0.01; mammary vein on d 180, P d 180 = 0.02) and TG (mammary vein on d 180, P d 180 < 0.01) were greater in the LP group than in the HP group. On d 180, the plasma concentration of TG in the caudal artery was higher in the LP group than in the HP group (P d 180 < 0.01). No significant differences in the plasma concentrations of BHBA and NEFA were found between the two groups (P persistency > 0.10).
Table 4. Plasma physiological and biochemical variables of dairy cows with high and low lactation persistency at d 0, 90 and 180 of the experiment1

Notes:
1 The data are expressed as the mean ± SEM, n = 12.
a,b Means without a common superscript differ significantly between the two groups at the same time point (P < 0.05).
2 TG, triglyceride; BHBA, β-hydroxybutyric acid; NEFA, non-esterified fatty acid.
3 HP, high lactation persistency; LP, low lactation persistency; d 0, day 0 of the experiment; d 90, day 90 of the experiment; d 180, day 180 of the experiment.
4 D, day; P, persistency; D × P, interaction between day and persistency.
Mammary tissue apoptosis and RNA-seq analysis
TUNEL staining of mammary gland tissue sections for cell apoptosis is given in Fig. 3. From the whole experiment period, the apoptosis of mammary cells in cows of HP (P d < 0.01) and LP (P d < 0.01) groups increased with the prolongation of lactation period. Moreover, the level of mammary cell apoptosis was significantly higher (P persistency < 0.01) in the LP group than in the HP cows throughout the entire trial. Furthermore, the mammary glands of LP cows had higher cellular apoptosis at d 0 (P d 0 = 0.05) in relative to those in HP group. Compared with HP cows, higher mammary apoptosis was observed in the LP cows at d 90 (P d 90 = 0.05) and d 180 (P d 180 < 0.01).

Figure 3. The levels of mammary apoptosis of dairy cows with high (HP) and low (LP) lactation persistency detected by the terminal deoxynucleotidyl transferase-mediated deoxyuridine triphosphate nick end labeling (TUNEL) assay. A Representative images of TUNEL staining. Green fluorescence reflects TUNEL-positive cells and blue fluorescence (DAPI) labels nuclei. Shown from top to bottom are at d 0, d 90 and d 180, with HP and LP included at each time point. B Quantification of TUNEL-positive cell apoptosis rate. Error bars indicate standard error of the mean. *P < 0.05, **P < 0.01.
The RNA-seq was performed to analyze the transcriptome difference in the mammary gland between HP and LP groups on d 0. Over 4.3 × 107 clean reads per sample were obtained after data filtering in all 14 mammary gland samples. The quality of the clean reads characterized by Q30 was more than 93%, and the GC content was in the range of 38–50% (Table S2). The following analysis was based on the mapped reads, and raw data were submitted to the National Center for Biotechnology Information database (BioProject PRJNA1049587). The principal component analysis was performed after preprocessing the raw data (Fig. 4A). The cutoff threshold of DEGs was |fold change| > 2 and P < 0.05. In total, 1135 DEGs were identified between the two groups (Table S3), of which 813 genes were upregulated and 322 genes were downregulated in the LP group compared with the HP group (Fig. 4B). KEGG pathway enrichment analysis showed that DEGs were assigned to 66 enriched KEGG pathways (P < 0.05) (Table S4), with enriched functions of the apoptosis pathway (P = 0.012) and the HIF-1 signaling pathway (P = 0.033, Fig. 4C).

Figure 4. The RNA-seq analysis of mammary tissues of dairy cows with high (HP) and low (LP) lactation persistency at d 0 of the experiment. A Principal component analysis (PCA) of HP and LP cows. B Volcano plot for mRNAs in HP vs. LP. The vertical dotted line delimits up-(red) and down-(blue) regulation (|log2-fold change| > 1, P < 0.05). C KEGG pathway enrichment analysis of differentially expressed genes (DEGs). Bubble charts represent the key enriched KEGG pathways. D Heat map of DEGs assigned to apoptosis and HIF-1 signaling pathway. Values represent the Z-scores of gene count. Red and blue colors represent up- and downregulation, respectively. E Network analysis of apoptosis and HIF-1 signaling pathway based on Pearson correlation. Red nodes represent genes in apoptosis pathway. Blue nodes represent genes in HIF-1 signaling pathway. The node size is proportional to between indegree. Red lines represent position correlations between nodes, and the blue represent negative correlations (|correlation| > 0.85 and P < 0.05). n = 7.
It was revealed that 16 genes significantly enriched in the apoptosis pathway (Fig. 4D). Expression of all of these 16 genes, except LOC784541, CSF2RB and GADD45G, was upregulated in the LP group compared with the HP group. Meanwhile, 12 genes were significantly enriched in the HIF-1 signaling pathway, 10 of them (except TFRC and ALDOC) were upregulated in the LP group compared with the HP group (Fig. 4D, Table S5 and S6). To better understand the potential causal effect of the HIF-1 signaling pathway on the apoptosis pathway, we conducted a network analysis of the interactions between genes, highlighting expression relevance between these genes from both KEGG pathways (cutoff at |correlation| > 0.85 and P < 0.05, Fig. 4E). The result showed that the expression of most genes in the two pathways was strongly correlated. Among them, FOS, a key gene involved in apoptosis, was closely connected with genes in the HIF-1 signaling pathway in the network.
In vitro study of hypoxia and cell apoptosis and oxidative stress
The levels of cell apoptosis and oxidative stress-related indicators in MAC-T cells under 1% and 11% oxygen concentrations are displayed in Fig. 5. Cell apoptosis rate (P < 0.01) increased significantly with hypoxic conditions compared to the 11% O2. The concentrations of ROS (P < 0.01) and MDA (P < 0.01) in culture medium were higher in 1% O2 than in 11% O2. Compared with cell culture in 11% O2, the levels of antioxidant-related indexes SOD (P < 0.01), GSH-Px (P < 0.01), T-AOC (P < 0.01) and T-NOS (P < 0.01) of cells in 1% O2 were lower.

Figure 5. The level of cell apoptosis and oxidative stress of MAC-T cells cultured under hypoxia (1% O2) and physiological oxygen concentration of dairy cows (11% O2) in vitro. A Cell apoptosis detected by Hoechst 33258 staining. On the left is the representative images of Hoechst 33258 staining and on the right is quantification of apoptotic cells. B–C The oxidative stress was assessed by intracellular reactive oxygen species (ROS, B) and malondialdehyde (MDA, C). D The antioxidant capacity was evaluated by superoxide dismutase (SOD), glutathione peroxidase (GSH-Px), total antioxidant capacity (T-AOC) and total nitric oxide synthase (T-NOS). n = 12. Error bars indicate standard error of the mean. **P < 0.01.
Discussion
Although lactation persistency is an important economic trait for dairy cows, the biological mechanism underlying lactation persistency difference among cows is still unclear. Physiological factors, such as hormone secretion and absorption of nutrients, play important role in the lactation characteristics of dairy cows (Gross Reference Gross2022). Although the role of hypoxia in multiple tissues is revealed, how oxygen metabolism influences mammary physiology remain to be investigated. In the present large-scaled animal study, we surprisingly observed that lactating cows with lower lactation persistency have higher HIF-1α during peak lactation period compared with cows with high lactation persistency, which is associated with elevated mammary gland apoptosis. Moreover, this study suggest that LP cows may be a relative hypoxic environment in the mammary gland tissue, which may induce oxidative stress, depressed glucose/lipid metabolic activities and enhanced cell apoptosis, leading to a rapid drop in milk yield.
In this study, cows with different lactation persistency had similar milk yield during peak lactation. The milk yield of the whole experiment changed following the normal progress of lactation, which was not affected by mammary gland biopsy procedures (Daley et al. Reference Daley, Dye, Bogers, Akers, Rodriguez, Cant, Doelman, Yoder, Kumar, Webster and Hanigan2018). A well-maintained functionality of the mammary gland depends greatly on the balance between cell proliferation (Capuco et al. Reference Capuco, Ellis, Hale, Long, Erdman, Zhao and Paape2003; Stefanon et al. Reference Stefanon, Colitti, Gabai, Knight and Wilde2002) and apoptosis (Yart et al. Reference Yart, Dessauge, Finot, Barbey, Marnet and Lollivier2012; Zheng et al. Reference Zheng, Ning, Dong, Zhao, Li, Fan, Li, Yu, Mrode and Liu2017). Although difference in milk yield between two groups initiated from d 105, TUNEL assay of the mammary gland showed that differences in cell apoptosis initiated from peak lactation, suggesting that management of mammary apoptosis toward high lactation persistency should be carried out from the early lactating stage. Intriguingly, LP cows had higher plasma HIF-1α concentration throughout the entire experimental period, with differences majorly attributed to altered HIF-1α level during peak lactation stage. According to previous studies, the accumulation of HIF-1α may be caused by different factors, such as lower oxygen levels due to high oxygen consumption (Liu et al. Reference Liu, Li and Xiong2022; Shao et al. Reference Shao, Wellman, Lounsbury and Zhao2014), alteration in health status induced by oxidative stress or inflammation (McGarry et al. Reference McGarry, Biniecka, Veale and Fearon2018) or increased energy metabolic requirements (Mattmiller et al. Reference Mattmiller, Corl, Gandy, Loor and Sordillo2011). The higher HIF-1α during peak lactation period may cause a sustained tissue apoptosis which has negative impacts on lactation performance during later lactating stages. We further verified the effect of hypoxia in mammary gland by using in vitro system, and results showed that higher apoptosis level of MAC-T cells was induced by hypoxia treatment, which further supported that higher HIF-1α may affect mammary apoptosis in vivo study (Nishimura et al. Reference Nishimura, Komiyama, Tasaki, Acosta and Okuda2008). Therefore, we speculated that higher HIF-1α in peak lactation cows may be the main effect inducing mammary apoptosis and the reduced lactation persistency.
Mammary transcriptome analysis was carried out to study the gene expression differences of two groups of cows on d 0. Our RNA-seq analysis showed major differences in expression of genes in HIF-1 signaling pathway and apoptosis pathway between HP and LP groups, which were consistent with our observations in HIF-1α, pO2 and cell apoptosis analyses in the mammary gland. Of particular interest, our network-based analysis suggested that the HIF-1α signaling pathway may play a predominant role in altering the expression of key functional genes associated with apoptosis. In addition, we found that an apoptosis pathway member, FOS, had a high degree in the network and was strongly linked with other genes of other activated pathways, especially HIF-1 signaling pathway. The FOS may form dimers with JUN proteins, activate activator protein-1 and start apoptosis process (Ameyar et al. Reference Ameyar, Wisniewska and Weitzman2003), as identified in different kinds of cells (Preston et al. Reference Preston, Lyon, Yin, Lang, Solomon, Annab, Srinivasan, Alcorta and Barrett1996; Smeyne et al. Reference Smeyne, Vendrell, Hayward, Baker, Miao, Schilling, Robertson, Curran and Morgan1993; Yadav et al. Reference Yadav, Kalra, Ganju and Singh2017). These observations provide potential targets in regulating the apoptotic status of the mammary gland to modulate lactation persistency of high-yielding dairy cows during peak lactation.
In addition, we noticed differences between two groups of cows in oxidative stress, glucose metabolism and lipid metabolism. We speculate that the differences observed in antioxidant capacity (MDA and GSH-Px) (Li et al. Reference Li, Yu, Xiao, Miao, Ji, Wang and Geng2017), NOS metabolism (iNOS and eNOS) (Jeffrey Man et al. Reference Jeffrey Man, Tsui and Marsden2014) and energy metabolism (glucose and TG) (Kierans and Taylor Reference Kierans and Taylor2021; Luo et al. Reference Luo, Li and Sang2023) may attributed to variation in apoptosis profiles between mammary of cows with different lactation persistency.
Conclusion
In this study, we found that although milk yield of cows with different lactation persistency showed difference only in late lactation stages, two groups of cows showed difference in cell apoptosis rate in the mammary gland, starting from peak lactation. The higher apoptosis rate in low lactation persistency cows was associated potentially with HIF-1α level in the mammary gland, which may be driven by related signaling pathways. Our findings identify a possible physiological factor responsible for low lactation persistency in dairy cows – hypoxia and its possible regulation ways of reducing lactation persistency by inducing mammary gland apoptosis. This provides a potential strategy related to hypoxia to improve lactation persistency of dairy cows in the future.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/anr.2024.5.
Data availability statement
All data generated or analyzed during this study are included in the published article.
Acknowledgements
The authors appreciate the staff of the Hangjiang Dairy Farm (Hangzhou, China) for their assistance in milking and caring for the animals. The members of the Institute of Dairy Science, Zhejiang University (Hangzhou, China) are acknowledged for their assistance with sampling and analysis of the samples.
Author contributions
ZZH, LL and YL performed the experiments. ZZH analyzed the data and wrote the original manuscript. JC, JXL and DMW contributed to the study design. JXL and DMW obtained funding and revised the manuscript. All authors read and approved the final manuscript.
Financial support
This work described in this article was financially supported by the National Natural Science Foundation of China (No. 31930107) and the China Agricultural (Dairy) Research System (CARS-36, Beijing). We also acknowledge the members of the Institute of Dairy Science Zhejiang University (Hangzhou, China) for their assistance when conducting these studies.
Conflict(s) of interest
The authors declare that they have no competing interests.
Ethical standards
This study was conducted in accordance with the Chinese guidelines for animal welfare and the experimental protocols for animal care approved by the Animal Care Committee of Zhejiang University (Hangzhou, Zhejiang, China) and was in accordance with the university’s guidelines for animal research.












