Introduction
Ariidae (Siluriformes) species, also known as marine catfish, are widespread in the tropical and subtropical continental shelves of the Atlantic, Indian and Pacific Oceans (Marceniuk et al., Reference Marceniuk, Menezes and Brito2012). Currently, a total of 156 Ariidae species belonging to 34 genera are known and ~20 species have been found to occur on the Atlantic coast of South America (Marceniuk et al., Reference Marceniuk, Menezes and Brito2012; Fricke et al., Reference Fricke, Eschmeyer and Van der Laan2022).
There are 77 species of Monogenoidea currently parasitizing Ariidae around the world; 68 Dactylogyridae (Chauhanellus Bychowsky & Nagibina, Reference Bychowsky and Nagibina1969, Hamatopeduncularia Yamaguti, 1953; Neotetraonchus Bravo-Hollis, 1968 and Susanlimocotyle Soares, Domingues & Adriano, 2021), seven Neocalceostomatidae (Fridericianella Brandes, 1894, Neocalceostoma Tripathi, 1959, Neocalceostomoides Kritsky, Mizelle & Bilqees, 1978 and Thysanotohaptor Kritsky, Shameem, Kumari, & Krishnaveni, 2012), and two Udonellidae (Udonella Johnson, 1835) (Lim et al., Reference Lim, Timofeeva and Gibson2001; Domingues et al., Reference Domingues, Soares and Watanabe2016; Illa et al., Reference Illa, Shameem, Serra, Melai, Mangam, Basuri, Petroni and Modeo2019; Soares et al., Reference Soares, Domingues and Adriano2021a, Reference Soares, Domingues and Adrianob; Soo & Tan, Reference Soo and Tan2021).
Chauhanellus, as amended by Lim (Reference Lim1994), is the second most species-rich genus, with 30 valid species, behind only Hamatopeduncularia, which comprises 32 species (Lim et al., Reference Lim, Timofeeva and Gibson2001; Soares et al., Reference Soares, Domingues and Adriano2021a; Soo & Tan, Reference Soo and Tan2021). Chauhanellus species possess a haptor with or usually without a digit-like extension; a dorsal anchor with or without spines at proximal base surface and slightly expanded outer roots; a ventral anchors with outer roots usually expanded and base of inner roots thickened; and a vaginal dextral opening with a sclerotized vaginal tube. Of the Chauhanellus species known to date, 28 have been described as infecting ariid marine catfish worldwide, while only two species have also been reported in non-ariid hosts (see table 1) (Lim et al., Reference Lim, Timofeeva and Gibson2001; Domingues & Fehlauer, Reference Domingues and Fehlauer2006; Domingues et al., Reference Domingues, Soares and Watanabe2016).
Table 1. Chauhanellus species from ariid hosts.

The new species described in this study is in boldface type.
a Formerly Arius falcarius
b Formerly Arius caelatus
c Formerly Tachysurus dussumieri
d Formerly Arius sagor
e Formerly Arius thalassinus
f Siluridae
g Formerly Arius macrocephalus
h Formerly Arius nenga
iBagridae
jFormerly Arius sinensis
k Formerly Arius graeffei
In South America, only six species of Chauhanellus have been described in Atlantic coast ariids (table 1) (Domingues & Fehlauer, Reference Domingues and Fehlauer2006; Domingues et al., Reference Domingues, Soares and Watanabe2016; Soares et al., Reference Soares, Domingues and Adriano2021a). However, recent studies indicate that the actual number of species in the region is probably much higher (Soares et al., Reference Soares, Adriano, Domingues and Balbuena2022).
During a study of monogenoids of ariid species captured from the Brazilian coast, a new species of Chauhanellus was found in the gills of Genidens barbus (Lacepède) and Genidens genidens (Cuvier) and is described based on morphological characters and partial 18S rDNA sequences. Supplementary taxonomic data from Chauhanellus velum Domingues, Soares & Watanabe, Reference Domingues, Soares and Watanabe2016, a parasite of Sciades couma (Valenciennes) (type-host) and new partial 18S rDNA sequences of Chauhanellus spp. from South American ariids, are also presented. Finally, new insights into Chauhanellus are provided based on morphology and molecular phylogenetic evidence.
Material and methods
Sample collection, morphological study and deposit of the helminths
Specimens of distinct ariid species were collected by local fishermen with trammel nets and hooks from four locations on the Brazilian coast between December 2017 to December 2019 under a License for the Collection of Zoological Material (SISBio No. 60666‒2 and Sisgen No. AD28DC2) (table 2). The gill arches were removed and placed in vials containing heated water (~65°C), following which each vial was vigorously shaken. The contents of each vial were examined in the laboratory under a dissecting microscope and helminths were removed from the gills or sediment using small probes. Helminths were fixed in either 4% formalin for morphological study or 96% ethanol for molecular characterization. Some specimens were stained with Gomori's trichrome and mounted in Damar gum to examine their internal soft structures, while others were mounted in Hoyer's medium (Humason, Reference Humason1979; Boeger & Vianna, Reference Boeger, Vianna and Thatcher2006) for the study of the sclerotized structures. Measurements, all in micrometres, were taken following the procedures of Mizelle & Klucka (Reference Mizelle and Klucka1953). Dimensions of organs and other structures represent the highest measurements in the dorso-ventral view; lengths of curved or bent structures (bars and accessory piece) represent the straight-line distances between the extreme ends; anchor length measurements followed Soares et al. (Reference Soares, Magalhães, Silva, Carneiro, Barbosa, Costa and Domingues2019); and the total length of the male copulatory organ (MCO) was measured using ImageJ (Rasband, Reference Rasband2022). Measurements are presented in micrometres as the mean followed by the range and the number (n) of specimens measured is shown in parentheses. Illustrations were prepared with a drawing tube attached to a Leica DM 2500 microscope with differential interference contrast and phase contrast optics. The soft structures were illustrated using pen and ink, while the hard structures were scanned and redrawn on a digitizing tablet using CorelDraw (Reference CorelDraw2014). Plates were also prepared in CorelDraw (Reference CorelDraw2014). Definitions of prevalence, mean intensity and mean abundance followed Bush et al. (Reference Bush, Lafferty, Lotz and Shostak1997). The Bray–Curtis similarity test (Bray & Curtis, Reference Bray and Curtis1957) was used to evaluate possible morphometric similarities between the specimens of Chauhanellus from S. couma (present study) and C. velum from other previously described hosts. Type specimens, vouchers and hologenophores (Pleijel et al., Reference Pleijel, Jondelius, Norlinder, Nygren, Oxelman, Schander, Sundberg and Thollesson2008) were deposited in the Invertebrate Collection of the Museu Paraense Emílio Goeldi (MPEG. PLA), Belém, Pará, Brazil, and the collection of Platyhelminthes of the Adão José Cardoso Museum of Zoology of the State University of Campinas (ZUEC PLA), São Paulo, Brazil, respectively, under No. (MPEG.PLA 000359–000384; ZUEC PLA 186-187). The vouchers of C. velum CHIOC 38262 a‒b, 38263 were examined for comparative purposes. Nomenclature for hosts followed Marceniuk et al. (Reference Marceniuk, Menezes and Brito2012). Details of the new taxa have been submitted to ZooBank following the International Code of Zoological Nomenclature (article 8.5 of the amended version) (International Commission on Zoological Nomenclature, 2012).
Table 2. Host species, locality (geographical coordinates), and monogenoid species detected in the present effort on each fish species.

N, number of host; PA, Pará; SP, São Paulo; RS, Rio Grande do Sul; BR, Brazil.
a Specimen subjected to molecular analysis, for which sequences of 18S rDNA were used for the phylogenetic analysis.
Molecular characterization of parasites
Each monogenoid specimen subjected to molecular analysis was divided using fine needles under a dissecting microscope. The anterior half of the body (without the MCO) was placed in a 1.5 ml microtube with 96% ethanol for genomic DNA extraction. The posterior part containing the haptoral complex and the MCO were flattened under coverslip pressure and mounted in Hoyer's solution for species identification. These fragments also served as vouchers (hologenophores). Genomic DNA was extracted using a Qiagen Dneasy® Blood and Tissue Kit, according to the manufacturer's protocol, with a final volume of 30 μl. DNA concentration was verified using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Massachusetts, USA) at 260 nm.
The partial 18S rDNA was amplified using a two-round polymerase chain reaction (PCR). In the first round, DNA was amplified with the primer pair WormA and WormB (Littlewood & Olson, Reference Littlewood, Olson, Littlewood and Bray2001). In the second round, for the semi-nested PCRs, the primer combinations were WormA and 1270R (Littlewood & Olson, Reference Littlewood, Olson, Littlewood and Bray2001) and 930F (Littlewood et al., Reference Littlewood, Waeschenbach and Nikolov2008) with WormB, which amplified two overlapping fragments of approximately 1179 base pairs (bp) and 1054 bp, respectively. PCRs were performed in a Matercycler® nexus (Eppendorff, Hamburg, Germany) with a final volume of 25 μl: 12.5 μl of DreamTaq Green PCR Master Mix (2×) (Thermo Scientific Wilmington, USA), following the manufacturer's recommendations, 0.5 mm of each primer, and 3 μl of the extracted DNA.
The PCR profile was performed using the cycling described in Soares et al. (Reference Soares, Domingues and Adriano2021a). The semi-nested PCRs were conducted with 1 μl of the product of the PCRs, diluted 1:1 in ultrapure water, applying the same cycling conditions. Amplicons were electrophoresed in 2% agarose gel in a TAE buffer (Tris 40 mm, acetic acid 20 mm, EDTA 1 mm) stained with SYBRsafe® (Invitrogen, Thermo Fisher Scientific, Massachusetts, USA) alongside a 1 kb Plus DNA Ladder (Invitrogen, Thermo Fisher Scientific, Massachusetts, USA) at 100 V for 30 min. The PCR products were purified using a QIAquick PCR Purification Kit (Qiagen, USA) and sequencing was carried out with the BigDye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems™) in a 3500 DNA sequencing analyser (Applied Biosystems, California, USA) at Helixxa Company (Paulínia, São Paulo, Brazil) or at the Human Genome Research Center, of the University of São Paulo (São Paulo, Brazil), using the same primers as used for rDNA amplification.
Alignment and phylogenetic inference
Contigs were edited using Sequencher 4.1.4 (Gene Codes, Ann Arbor, MI) and deposited in GenBank under the accession numbers listed in table 3. Standard nucleotide Basic Local Alignment Search Tool searches were then conducted (Altschul et al., Reference Altschul, Madden, Schaffer, Zhang, Zhang, Miller and Lipman1997) to verify the similarity of the newly obtained sequences from the present study with other sequences of monogenoids in the United States National Center for Biotechnology Information (NCBI) BioSystems database (Geer et al., Reference Geer, Marchler-Bauer, Geer, Han, He, He, Liu, Shi and Bryant2009). Alignment of the 18S rDNA was generated using MUSCLE implemented in Geneious version 7.1.3 (Kearse et al., Reference Kearse, Moir and Wilson2012). A total of 34 partial sequences of the 18S rDNA of species belonging to the Dactylogyridea order published in the NCBI BioSystems database (Geer et al., Reference Geer, Marchler-Bauer, Geer, Han, He, He, Liu, Shi and Bryant2009), along with two of the Monocotylidea order (used as the outgroup) were retrieved from GenBank (see table 3) and aligned with five newly generated sequences of Chauhanellus spp. from ariids from the Brazilian coast (Chauhanellus riograndinensis n. sp., Chauhanellus neotropicalis Domingues & Fehlauer, Reference Domingues and Fehlauer2006, Chauhanellus hamatopeduncularoideum Domingues, Soares & Watanabe, Reference Domingues, Soares and Watanabe2016, Chauhanellus hypenocleithrum Domingues, Soares & Watanabe, Reference Domingues, Soares and Watanabe2016, and C. velum). Despite the availability of several partial 18S rDNA sequences, we restricted our analysis to sequences of monogenoids >1600 bp to attain the highest number of variable and phylogenetically informative sites. Forty-one sequences (1647–2200 bp long) were aligned, and the extremes were trimmed, leaving an alignment of 1777 bp in length. The model of evolution was selected by JModelTest 2.1.1 (Darriba et al., Reference Darriba, Taboada, Doallo and Posada2012) using the Akaike information criterion. Phylogenetic analyses were performed using the maximum likelihood (ML) and Bayesian inference (BI) methods. ML was performed in PhyML 3.0, implemented via a web server (http://www.atgc‒montpellier.fr/phyml/) (Guindon et al., Reference Guindon, Dufayard, Lefort, Anisimova, Hordijk and Gascuel2010), with topology assessed by bootstrapping with 1000 replicates, applying the GTR + I + G model. BI was performed using MrBayes v.3.2.6 (Ronquist et al., Reference Ronquist, Teslenko and Mark2012), under the same model, with posterior probabilities estimated from 5 × 105 generations with two independent runs of four simultaneous Markov chain Monte Carlo (MCMC) algorithms, sufficient to keep the average standard deviation below 0.001 and the effective sample size (>200) on Tracer v1.7 (Rambaut et al., Reference Rambaut, Drummond, Xie, Baele and Suchard2018). The MCMC algorithms with the 1000th tree saved and diagnostics for every 1000th generation with burn-in periods, were set to the first 25,000 generations. Trees were visualized using FigTree 1.3.1 (Rambaut, Reference Rambaut2022) and figures prepared using CorelDraw (Reference CorelDraw2014). Genetic divergence was determined using the P-distance model matrix in MEGA version 7 (Kumar et al., Reference Kumar, Stecher and Tamura2016). Gaps and missing data were deleted.
Table 3. List of monogenoids included in the phylogenetic analyses, providing host species data, locality, GenBank ID and references. Data for the present study are highlighted with boldface type.

a Euryhaliotrematoides was placed in subjective synonymy with Euryhaliotrema (Kritsky, Reference Kritsky2012).
b Species used as outgroups.
Results
A total of 100% of the 10 host specimens of Amphiarius rugispinis (Valenciennes), 100% of the fourteen Aspistor quadriscutis (Valenciennes), five (16%) of the 31 host specimens of G. barbus, seven (42%) of the seventeen G. genidens, three (50%) of the six Sciades couma and 100% of the nine S. proops (Valenciennes) examined were infected with monogenoids (table 2).
The morphological, morphometric and partial 18S rDNA data endorsing the proposition of the new taxon, the supplementary taxonomic of C. velum and insights into Chauhanellus are presented below.
Taxonomic acts
Taxonomic summary
Class Monogenoidea Bychowsky, 1937
Subclass Polyonchoinea Bychowsky, 1937
Order Dactylogyridea Bychowsky, 1937
Dactylogyridae Bychowsky, 1933
Chauhanellus Bychowsky & Nagibina, Reference Bychowsky and Nagibina1969
Chauhanellus riograndinensis n. sp. (fig. 1)
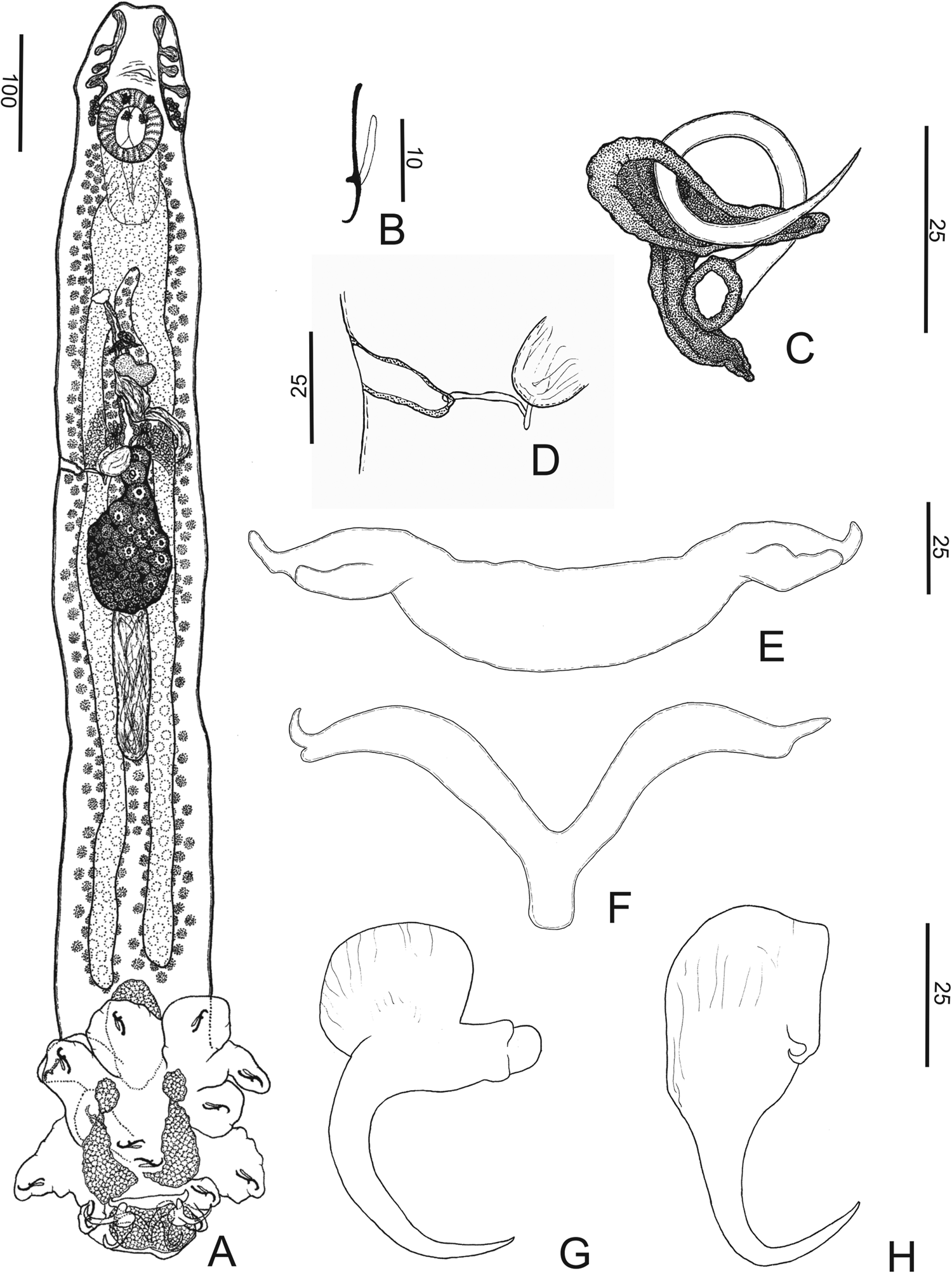
Fig. 1. Chauhanellus riograndinensis n. sp. (a) Holotype whole-mount, ventral; (b) hook; (c) copulatory complex; (d) vagina; (e) ventral bar; (f) dorsal bar; (g) ventral anchor; (h) dorsal anchor. Scale bars: (a) 100 μm; (b) 10 μm; (c–h) 25 μm.
Synonym. Chauhanellus sp. (Soares et al., Reference Soares, Adriano, Domingues and Balbuena2022)
Type-host. Genidens barbus (Lacepède), (Siluriformes, Ariidae).
Site of infection. Gills.
Type locality. Estuary of Patos Lagoon, Municipality of Rio Grande, Rio Grande do Sul State, Brazil (32° 08′ 05.7″ S; 52° 06′ 11.2″ W).
Other records. Genidens genidens (Cuvier), Ariidae (prevalence: 42% of 17 hosts; mean intensity: 47; mean abundance: 1.1), Estuary of Patos Lagoon, Municipality of Rio Grande, Rio Grande do Sul State, Brazil (32° 08′ 05.7″ S; 52° 06′ 11.2″ W).
Prevalence. 16% of 31 hosts examined.
Mean intensity. 32 parasites per infected host.
Mean abundance. two parasites per host.
Specimens deposited. Holotype, MPEG.PLA 000359; paratypes, MPEG.PLA 000360‒000372; vouchers, MPEG.PLA 000373‒000379; hologenophore, ZUEC PLA 187.
Representative DNA sequence. 1689 bp long partial sequence of the 18S rDNA gene of one parasite isolates (GenBank accession number, OQ517175).
Etymology: The specific name refers to the municipality of Rio Grande, Rio Grande do Sul State, Brazil, where the type host was collected.
Number of ZooBank. 7E70DF71-791A-4333-B6B7-7989BB6F4EA1
Comparative measurements. table 4
Table 4. Comparative measurements (in μm) of specimens of Chauhanellus riograndinensis n. sp. from Genidens barbus and Genidens genidens from Rio Grande, Rio Grande do Sul, Brazil.
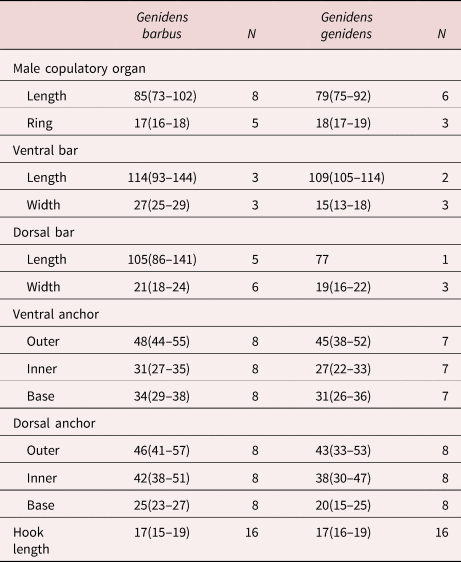
N, number of parasite.
Description. (Based on fourteen specimens, eight mounted in Hoyer's medium and six stained with Gomori's trichrome). Body fusiform, total length excluding haptor 863 (743–972; n = 6), total width at the level of germarium 141 (126–168; n = 6) (fig. 1a). Cephalic margin tapered; poorly developed terminal lobes; four bilateral pairs of head organs with rod-shaped secretion; cephalic glands unicellular, lateral to pharynx. Eyes four, equidistant; accessory chromatic granules absent. Mouth subterminal, midventral, prepharyngeal; pharynx comprising muscular, glandular bulb, spherical, 72 (63–85; n = 6) long, 71 (56–88; n = 6) wide. Oesophagus elongate; two intestinal caeca, non-confluent posteriorly, lacking diverticula. Common genital pore midventral near level of intestinal bifurcation; genital atrium muscular, unarmed. Gonads intercaecal, testis post-germarial, dorsal to germarium. Testis bacilliform, 120 (109‒134; n = 6) long, 52 (39‒65; n = 6) wide. Vas deferens looping left of intestinal caecum; seminal vesicle a dilatation of vas deferens, sigmoid. Single saculiform prostatic reservoir, lying posterior to the base of MCO. Copulatory complex comprising MCO, accessory piece; MCO, sclerotized, coiled tubular shaft of around one counterclockwise ring, base with a sclerotized cap, 85 (73–102; n = 8) long, tapered distal region. Accessory piece sclerotized, T-shaped, proximally spatulate, guarding termination of MCO; non-articulated with MCO (fig. 1c). Germarium piriform, 108 (79‒164; n = 6) long, 69 (48‒92; n = 6) wide. Uterus delicate. Vagina comprises a vaginal aperture with dextroventral opening, submarginal; vaginal vestibule cup-shaped, slightly sclerotized; narrow vaginal canal, a loop at the distal portion before entering the seminal receptacle (fig. 1d). Seminal receptacle ovoid. Mehlis’ glands, bilateral to the uterus. Eggs, ootype not observed. Vitellaria coextensive with intestinal caeca; transverse vitelline duct anterior to seminal receptacle, dorsal to intestinal caeca. Haptor digitiform, 242 (213‒273; n = 4) long, 222 (180‒242; n = 4) wide, with four haptoral glands (two ventral and two mid-dorsal). Anchors dissimilar. Ventral anchor, outer 48 (44–55; n = 8) long, inner 31 (27–35; n = 8) long; base 34 (29–38; n = 8) long; with divergent roots; truncate superficial root; expanded deep root, evenly curved elongate shaft, slightly straight short point extending at the level of tip of the superficial root (fig. 1g). Dorsal anchor, outer 46 (41–57; n = 8) long, inner 42 (38–51; n = 8) long; base 25 (23–27; n = 8) long; robust, with inconspicuous roots, slightly curved short shaft, elongate point extending well past the level of the tip of the inner base; anchor spine blunt (fig. 1h). Ventral bar, 114 (93‒144; n = 3) long, 27 (25‒29; n = 3) wide, broadly open U-shaped rod with bifid ends for articulation with ventral anchor (fig. 1e). Dorsal bar, 105 (86–141; n = 5) long, 21 (18–24; n = 6) wide, V-shaped, with bifurcation on both ends, acute anterior protuberance, rounded posterior protuberance; elongated posteromedial process (fig. 1f). Hooks similar in shape, 17 (15–19; n = 16) long, shank elongated, without inflation, erect thumb, lightly curved long shaft and delicate point (fig. 1b). Filamentous hook loop comprising 80% of the length of the shank.
Remarks
The new species seems to be closely related to Chauhanellus susamlimae Domingues, Soares & Watanabe, Reference Domingues, Soares and Watanabe2016, by having a digitate haptor; a ventral anchor with divergent roots, a truncate superficial root and an expanded deep root; evenly curved shaft; a dorsal anchor, robust, with inconspicuous roots; slightly curved shaft; elongated point; and anchor spine blunt. However, C. riograndinensis n. sp. can be easily distinguished from this species because the new species possesses MCO, a coiled tubular shaft of around one counterclockwise ring, base with a sclerotized cap, MCO with tapered distal region and accessory piece, T-shaped, proximally spatulate, guarding termination of MCO (MCO, a sclerotized tube, sigmoid, and accessory piece comprising an elongated sheath in C. susamlimae) and vagina sclerotized (a muscular vagina [unsclerotized] in C. susamlimae).
Chauhanellus velum Domingues, Soares & Watanabe, Reference Domingues, Soares and Watanabe2016 (fig. 2)

Fig. 2. Chauhanellus velum Domingues, Soares & Watanabe, Reference Domingues, Soares and Watanabe2016. (a) Voucher specimen whole-mount, ventral; (b) copulatory complex; (c) hook; (d) ventral bar; (e) dorsal bar; (f) ventral anchor; (g) dorsal anchor. Scale bars: (a) 100 μm; (c) 10 μm; (b) 25 μm; (d–g) 50 μm.
Type-host. Sciades couma (Valenciennes) (Siluriformes, Ariidae).
Site of infection. Internal borders of the primary lamellae of the gills.
Type-locality. Fish market, Municipality of Bragança, State of Pará, Brazil.
Other records. Sciades couma, Caratateua, Municipality of Bragança, State of Pará, Brazil (1° 59′ 41.91″ S, 46° 43′ 21.385″ W) (present study); Sciades herzbergii, Caratateua, Municipality of Bragança, State of Pará, Brazil (Soares et al., Reference Soares, Domingues and Adriano2021a); Sciades herzbergii, Furo da Ostra, municipality of Curuça, State of Pará, Brazil; Sciades passany, Fish market, Municipality of Bragança, State of Pará, Brazil (Domingues et al., Reference Domingues, Soares and Watanabe2016).
Prevalence. 50% of six hosts examined.
Specimens deposited. Vouchers five (MPEG.PLA 000380‒000384).
Specimens studied. Vouchers of C. velum CHIOC 38262 a‒b, 38263.
Representative DNA sequence. 1654 bp long partial sequence of the 18S rDNA gene of one parasite isolates (GenBank accession number, OQ511558).
Comparative measurements. table 5
Table 5. Comparative measurements (in μm) of specimens of Chauhanellus velum Domingues, Soares & Watanabe Reference Domingues, Soares and Watanabe2016 from Sciades couma, Sciades herzbergii and Sciades passany from different ariids’ hosts.

N, number of parasite.
a Present study.
b Domingues et al. (Reference Domingues, Soares and Watanabe2016).
Redescription. (Based on five specimens, two mounted in Hoyer's medium and three stained with Gomori's trichrome). Body fusiform, total length excluding haptor 898 (766–1143; n = 3), total width at the level of germarium 178 (148–232; n = 3) (fig. 2a). Cephalic margin tapered; poorly developed terminal lobes; four bilateral pairs of head organs with rod-shaped secretion; cephalic glands not observed. Eyes four, equidistant; accessory chromatic granules absent. Mouth subterminal, midventral; pharynx subspherical, 82 (70–100; n = 3) long, 69 (57–79; n = 3) wide. Oesophagus short; two intestinal caeca, non-confluent posteriorly, lacking diverticula. Common genital pore midventral, anterior to the copulatory complex near the pharynx; genital atrium muscular, unarmed. Gonads intercaecal, testis post-germarial, dorsal to germarium. Testis subspherical, 83 (72‒93; n = 2) long, 72 (70‒75; n = 2) wide. Vas deferens looping left of intestinal caecum; seminal vesicle a dilatation of vas deferens, sigmoid. Single spherical prostatic reservoir, lying posterior to the base of MCO. Copulatory complex comprising MCO, accessory piece; MCO sclerotized, coiled tubular shaft of around one counterclockwise ring, base with an expanded sclerotized cap, 103 (85–113; n = 4) long, MCO acute distal region. Accessory piece sclerotized, comprising two regions, proximal region with three small projections (one projection serrated on the inner margin) and duct throughout the MCO pass, distal region an elongated sheath; non-articulated with MCO (fig. 2b). Germarium piriform, 121 (115‒130; n = 3) long, 64 (50‒81; n = 3) wide. Uterus delicate. Vagina comprises vaginal aperture with dextroventral opening, submarginal; vaginal vestibule with soft tissue; vaginal canal, short, sclerotized, knob shape, before entering the seminal receptacle. Seminal receptacle ovoid. Mehlis’ glands, bilateral to the uterus. Eggs, ootype not observed. Vitellaria coextensive with intestinal caeca. Haptor subcircular, velum-like, 314 (232‒383; n = 3) long, 273 (199‒331; n = 3) wide, with four haptoral glands. Anchors dissimilar. Ventral anchor, outer 47 (39–54; n = 4) long, inner 44 (38–39; n = 4) long; base 39 (37–43; n = 4) long; with divergent roots; truncate superficial root; expanded deep root, mildly curved elongate shaft, slightly curved short point extending at the level of the tip of the superficial root (fig. 2f). Dorsal anchor, outer 44 (36–49; n = 3) long, inner 32 (31–33; n = 3) long; base 33 (28–37; n = 3) long; robust, with inconspicuous roots, expanded margin, slightly curved short shaft, elongate point extending well past the level of the tip of the inner base; anchor spine blunt (fig. 2g). Ventral bar 53 (42‒67; n = 5) long, 12 (10‒13; n = 4) wide, curved in the posterior direction, with bifid ends for articulation with ventral anchor (fig. 2d). Dorsal bar 53 (38–76; n = 5) long, 10 (7–14; n = 5) wide, slightly straight shaped, with rounded ends; mid-posteromedial process (fig. 2e). Hooks similar in shape, 18 (17–19; n = 5) long, shank elongated, without inflation, erect thumb, evenly curved shaft point (fig. 2c). Filamentous hook loop comprising 80% of the length of the shank.
Remarks
A comparative analysis of the vouchers of C. velum (CHIOC 38262 a‒b, 38263), provided by Domingues et al. (Reference Domingues, Soares and Watanabe2016), and specimens of Chauhanellus from S. couma, Caratateua, Municipality of Bragança, in Pará, Brazil, indicated that they are conspecific, mainly because they both share the same morphology of the copulatory complex, haptor velum-like, bars and anchors (fig. 2) (see also molecular data results). Moreover, the Bray–Curtis morphometric analysis of the morphological structures of C. velum from S. couma (present study) and S. herzbergii, S. couma and S. passany (table 5) identified a similarity of 88–95% across the specimens from each host (fig. 3; Online supplementary table S1). In addition, we provide supplementary morphological data and new illustrations of C. velum (fig. 2).

Fig. 3. Morphometric similarity dendrogram by Bray–Curtis method for Chauhanellus velum from different hosts and the new species described.
The ventral anchor observed in the specimens in the present study possesses a truncate superficial root and an expanded deep root (fig. 2f). This characteristic was also observed in the vouchers examined (CHIOC 38262 a‒b, 38263); however, not as clearly as in the specimens of the present study, as it appears to be compressed by the coverslip sheet, making its definition difficult. It is true that the species has a truncate superficial root, although it was not clear in the drawing from the original description (Domingues et al., Reference Domingues, Soares and Watanabe2016, p. 312, fig. 27). The compression of this structure by the coverslip sheet may have caused a misinterpretation by Domingues et al. (Reference Domingues, Soares and Watanabe2016). In addition, with respect to the intestinal caeca, vas deferens, genital pore, genital atrium, seminal vesicle, seminal receptacle, uterus and Mehlis’ gland, none of which were observed in the original description of the species, the present study provides a better definition of these structures (see above).
Molecular data
The sequencing of the partial 18S rDNA of C. riograndinensis n. sp. was 1689 bp in length. Beyond this, four new partial 18S rDNA sequences were obtained for four other species of Chauhanellus (C. neotropicalis from A. quadriscutis – 1699 bp long, C. hamatopeduncularoideum from A. rugispinis [type-host] – 1611 bp long, C. hypenocleithrum from S. proops [type-host] – 1653 bp long and C. velum from S. couma [type-host] –1654 bp long) (table 2).
The genetic divergence between Chauhanellus species and monogenoid species from Siluriformes was compared, varying from 3.2 to 11.8% (table 6). Interspecific divergence within Chauhanellus ranged from 0.3 to 4.6% (7‒189 bp). The genetic divergence among C. riograndinensis n. sp. and other Chauhanellus species was between 2.3 and 4.3% (39‒77 pb). The divergence between C. riograndinensis n. sp. and the most similar morphological species, C. susamlimae, was 2.2% (39 pb). The smallest interspecific distance was observed between C. boegeri Domingues & Fehlauer, Reference Domingues and Fehlauer2006 and C. neotropicalis at only 0.3% (7 bp), while C. velum was the most genetically distant species of Chauhanellus (4.6%). There was no intraspecific divergence between sequences of C. velum from the distinct hosts S. herzbergii and S. couma.
Table 6. Pairwise genetic identities of 18S rDNA sequences selected from Dactylogyridae species from Siluriformes adjusted for missing data.

The upper triangular matrix shows the number of differences in nucleotides and the lower triangular matrix shows the differences in terms of percentage of nucleotides.
Sequences obtained in the present work are shown in in boldface type.
a In Sciades herzbergii.
b In Sciades couma.
Phylogenetic evidence
The ML and BI phylogenetic analyses based on the 18S rDNA gene converged with similar topologies, and only the BI tree was presented, with the statistical support of both methods (fig. 4). Monogenoid species from Siluriformes fish arose in clade S, with highly supportive nodes in both ML and BI analyses, and were further divided into three subclades, S1, S2 and S3 (fig. 4). S1 exclusively comprises parasites of freshwater catfish from the Oriental region: Mizellus longicirrus (Tripathi, 1959) from Siluridae, Bychowskyella spp. from Heteropneustidae and Clariidae and Thaparocleidus spp. from Siluridae. S2 clustered exclusively parasites of marine catfish (Ariidae) from South America and the Oriental region: Susanlimocotyle narina from S. herzbergii arises forming a strongly supported lineage closely related to Hamatopeduncularia spp. (Hamatopeduncularia arii Yamaguti, 1953, Hamatopeduncularia bifida Illa, Shameem, Serra, Melai, Mangam, Basuri, Petroni & Modeo, Reference Illa, Shameem, Serra, Melai, Mangam, Basuri, Petroni and Modeo2019, Hamatopeduncularia elongata Lim,1996, Hamatopeduncularia thalassini Bychowsky & Nagibina, 1968 [all from Arius jella Day] and Hamatopeduncularia madhaviae Illa, Shameem, Serra, Melai, Mangam, Basuri, Petroni & Modeo, Reference Illa, Shameem, Serra, Melai, Mangam, Basuri, Petroni and Modeo2019, from Plicofollis dussumieri [Valenciennes]) from Oriental ariids (fig. 4).
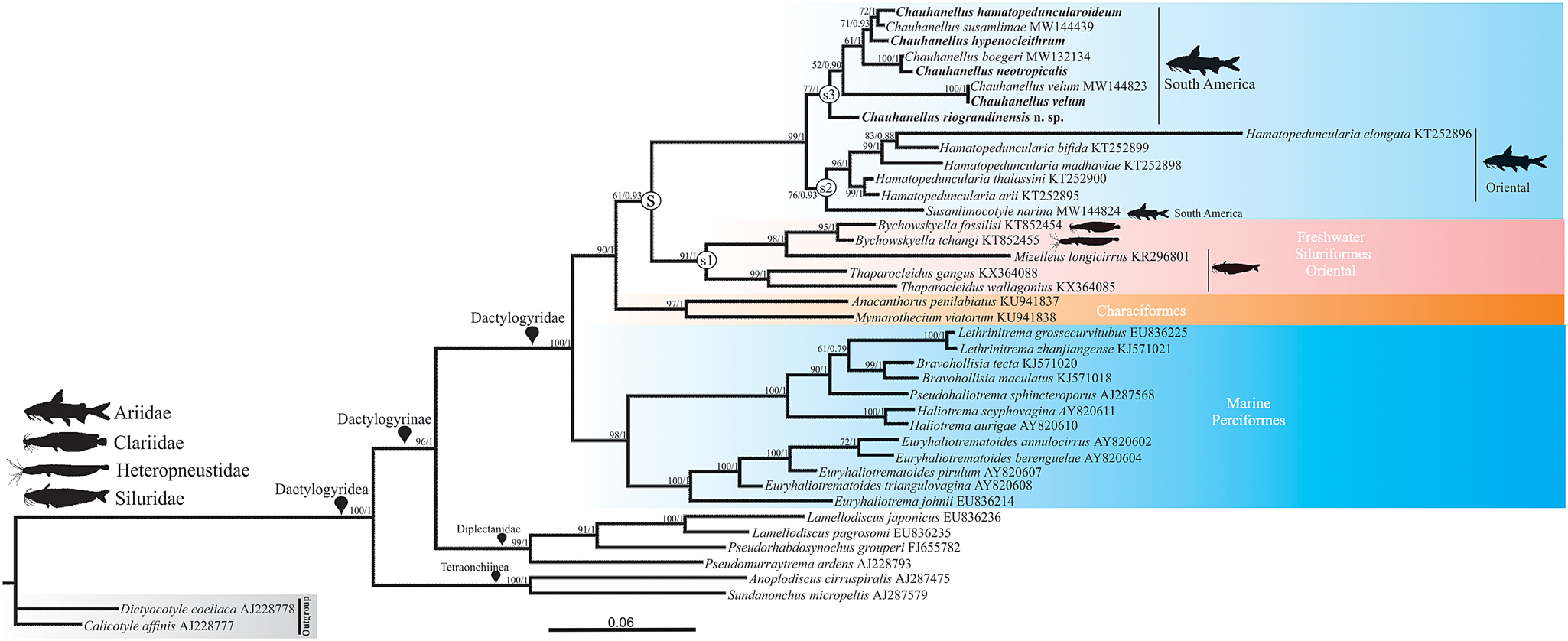
Fig. 4 Molecular phylogeny of the Dactylogyridea estimated by Bayesian inference using partial sequences of the 18S rDNA gene (1777 base pairs long). Species newly sequenced for the present study are in boldface type. Species name precedes the GenBank sequence ID. Maximum likelihood bootstrap support values and posterior probabilities are given above the branches (bootstrap values <50 and posterior probabilities <0.70 not reported).
Subclade S3 arises in a sister position to clade S2 and comprises Chauhanellus species that are exclusive parasites of marine catfish (Ariidae) from South America: C. hamatopeduncularoideum from A. rugispinis, and C. susamlimae from S. herzbergii, appearing as the derived species in a sister position to C. hypenocleithrum from S. proops. In turn, with strong support, this lineage appears as sister to the one composed of C. boegeri from S. herzbergii and C. neotropicalis from A. quadriscutis. Chauhanellus riograndinensis n. sp., a parasite of G. barbus and G. genidens, and C. velum from S. couma and S. herzbergii, arose as early divergent Chauhanellus species.
Discussion
Several genera have been proposed for the monogenoids of ariid fishes worldwide (Soares et al., Reference Soares, Domingues and Adriano2021a, Reference Soares, Domingues and Adrianob). Fridericianella, Neocalceostoma, Neocalceostomoides and Thysanotohaptor are harboured in the Neocalceostomatidae and Udonella in Udonellidae (Soares et al., Reference Soares, Domingues and Adriano2021a, Reference Soares, Domingues and Adrianob). The four others, Chauhanellus, Hamatopeduncularia, Neotetraonchus and Susanlimocotyle belong to the Dactylogyridae (Soares et al., Reference Soares, Domingues and Adriano2021a).
Of the six valid Chauhanellus species so far reported to be infesting South American ariid catfishes (table 1), only three have available sequences (Soares et al., Reference Soares, Domingues and Adriano2021a). In the present study, we sequenced all known South American Chauhanellus species, as well as the new species described here.
Only two prior reports exist of Chauhanellus species parasitizing G. barbus and G. genidens from the Brazilian coast: C. boegeri on G. barbus and G. genidens from Guaratuba, Paraná, Brazil (Domingues & Fehlauer, Reference Domingues and Fehlauer2006); and an undetermined species of Chauhanellus found on G. barbus and G. genidens from the south of Brazil (Soares et al., Reference Soares, Adriano, Domingues and Balbuena2022). Nevertheless, a Chauhanellus species reported but not described by Soares et al., Reference Soares, Adriano, Domingues and Balbuena2022 is formally described here.
Chauhanellus was proposed by Bychowsky & Nagibina (Reference Bychowsky and Nagibina1969) to accommodate two new species, C. oculatus Bychowsky & Nagibina, Reference Bychowsky and Nagibina1969 and C. flexiosus Bychowsky & Nagibina, Reference Bychowsky and Nagibina1969, from ariids in the south of China, as well as to transfer Ancyrocephalus alatus Chauhan, 1945 to Chauhanellus as C. alatus (Chauhan, 1945). The genus was proposed as closely related to Hamatopeduncularia (Bychowsky & Nagibina, Reference Bychowsky and Nagibina1969), differing from the latter by the absence of digitation on the haptor. Later, Lim (Reference Lim1994) proposed an amended diagnosis to the genus to accommodate some species that possess a haptor with or usually without digit-like extension and a dorsal anchor with or without spines at the proximal base surface, which are characteristics also found in Hamatopeduncularia (see Lim et al., Reference Lim, Timofeeva and Gibson2001). Here, C. riograndinensis n. sp. is described as possessing a combination of Chauhanellus-like characteristics (i.e. expanded deep roots on the ventral anchors, spine on the dorsal anchor, dumbbell-shaped ventral bar and dorsal bar with posteromedial process) and Hamatopeduncularia-like characteristics (i.e. haptoral digits).
Our analyses based on partial sequences of the 18S rDNA gene revealed phylogenetic support for the validity of C. riograndinensis n. sp., suggesting that this species is an early divergent Chauhanellus species from South America (fig. 4). Our results, based on morphological, morphometric and molecular data, also suggest that the specimens of Chauhanellus from S. couma reported in the present study and C. velum from S. herzbergii, S. couma and S. passany reported by Domingues et al. (Reference Domingues, Soares and Watanabe2016) and Soares et al. (Reference Soares, Domingues and Adriano2021a) are conspecific (figs 3 and 4, tables 5 and 6, Online supplementary table S1).
To date, there are ten known monogenoid species that possess a combination of the characteristics of Chauhanellus and Hamatopeduncularia (Kearn & Whittington, Reference Kearn and Whittington1994; Lim, Reference Lim1994, Reference Lim1996; Lim et al., Reference Lim, Timofeeva and Gibson2001; Domingues et al., Reference Domingues, Soares and Watanabe2016). For example, Chauhanellus intermedius Lim, Reference Lim1994, C. digitalis Lim, Reference Lim1994, C. aspinous Lim, Reference Lim1994, C. pedunculatus Paperna,1977, C. hamatopeduncularoideum and C. susamlimae possess characteristics found in Chauhanellus (i.e. anchor with root expanded into wings and ventral bar with protuberances at each end) and Hamatopeduncularia (i.e. haptoral digits and absence of spines on the dorsal anchor); while Hamatopeduncularia arii, H. thalassani, H. pulchra Bychowsky & Nagibina, Reference Bychowsky and Nagibina1969 and H. pearsoni Kearn & Whittington, Reference Kearn and Whittington1994, exhibit characteristics found in Hamatopeduncularia (i.e. haptoral digits and absence of spines on the dorsal anchor) and Chauhanellus (i.e. anchor with root expanded into wings and ventral bar with protuberances at each end). The sharing of these morphological characteristics between Chauhanellus and Hamatopeduncularia has led some authors to raise the question of synonymy (Kearn & Whittington, Reference Kearn and Whittington1994; Lim, Reference Lim1994, Reference Lim1996; Lim et al., Reference Lim, Timofeeva and Gibson2001; Domingues et al., Reference Domingues, Soares and Watanabe2016; Soares et al., Reference Soares, Domingues and Adriano2021a). However, Soares et al. (Reference Soares, Domingues and Adriano2021a), based on phylogenetic analyses (using the partial 18S rDNA sequences >1700 bp) to support the validity of both genera and suggested that a morphological reevaluation of Chauhanellus and Hamatopeduncularia is required.
Recent studies based on molecular data (partial sequences of 18S rDNA and 28S rDNA < 900 bp) have shown that some species of Chauhanellus appear nested with other species of Hamatopeduncularia in their phylogenetic analyses, suggesting the non-monophyly of each genus (Illa et al., Reference Illa, Shameem, Serra, Melai, Mangam, Basuri, Petroni and Modeo2019; Soo & Tan, Reference Soo and Tan2021). However, the use of inappropriate sequences (e.g. short or unpublished sequences) may have contributed to these results.
The use of short sequences in phylogenetic analysis limits comparison, especially in relation to closely related taxa, as it restricts the number of variable and phylogenetically informative sites (Littlewood & Olson, Reference Littlewood, Olson, Littlewood and Bray2001). In addition, small changes in alignment can have major effects on phylogenetic reconstruction (Winnepenninckx & Backeljau, Reference Winnepenninckx and Backeljau1996), and the effects of missing data can have an undesirable influence on resulting trees (Barriel, Reference Barriel1994; Wilkinson, Reference Wilkinson1995). We suggest that future studies that seek to test the monophyly of Hamatopeduncularia and Chauhanellus use long sequences, molecular markers from different regions and include a wider range of taxa, including the type species of each genus (H. arii and C. oculatus) and representatives of New-World and Old-World lineages sensu Soares et al., Reference Soares, Domingues and Adriano2021a.
Despite the fact that G. barbus from Cananéia, São Paulo, Brazil (present study) and G. barbus and G. genidens from Guaratuba, Paraná state, Brazil (Domingues & Fehlauer, Reference Domingues and Fehlauer2006) had been examined, the current new species was not found in these locations. On the other hand, C. boegeri from Guaratuba, Paraná, Brazil, was found in G. barbus and G. genidens obtained from the current study area (i.e. Cananéia, São Paulo, south-eastern Brazil and Rio Grande, Rio Grande do Sul state, southern Brazil), as well as on S. couma from Caratateua, Pará, northern Brazil (table 2). According to Lim et al. (Reference Lim, Timofeeva and Gibson2001), the presence of different species of Chauhanellus on the same host species in different biogeographical regions (e.g. Arius maculatus (Thunberg) from Hainan, China, and from the western coast of Peninsular Malaysia) suggests that habitat differences may affect the presence or absence of certain parasite species. However, the limitations of insufficient numbers of host samples and potentially misidentified host species should also be considered.
Conclusion
The present study provides morphological and molecular characterization (partial 18S rDNA) of a new species of Chauhanellus and new 18S rDNA sequences of Chauhanellus spp. from South American ariids. Our results showed that C. riograndinensis n. sp. and C. velum represent two early divergent lineages within Chauhanellus from South America. Moreover, the confirmation of the conspecificity of Chauhanellus specimens from S. couma (present study) and C. velum from S. couma, S. herzbergii, and S. passany reported by other authors emphasizes the need for an integrative taxonomic approach to ensure accurate delimitation of monogenoid species. Finally, we suggest one way for future studies that seek to test the monophyly of Chauhanellus and Hamatopeduncularia.
Supplementary material
To view supplementary material for this article, please visit https://doi.org/10.1017/S0022149X23000135.
Acknowledgement
We thank Marcelo Knoff, Coleção Helmintológica do Instituto Oswaldo Cruz (Brazil), who allowed us access to specimens under his care.
Financial support
The present study was partly supported by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (the Coordination for the Improvement of Higher Education Personnel) – Brazil (CAPES) – Finance Code 001. G.B. Soares was supported by a doctoral scholarship #2017/17531-0, São Paulo Research Foundation (FAPESP). M. V. Domingues and E. A. Adriano received research productivity grants from the Brazilian Fostering Agency CNPq (respectively, grant #309896/2019-3 and #301886/2016-4).
Conflicts of interest
None.
Ethical standards
Permission to collect the hosts was given by the Brazilian Institute of Environment and Renewable Natural Resources (IBAMA) under a License for the Collection of Zoological Material (SISBio No. 60666–2) and for access to genetic heritage (Sisgen No. AD28DC2).












