Introduction
Data acquisition rates in fluorescence microscopy are exploding due to the increasing size and sensitivity of detectors, brightness and variety of available fluorophores, and complexity of the experiments and imaging equipment. An increasing number of laboratories are now performing complex imaging experiments that rapidly generate gigabytes and even terabytes of data, but the practice of data storage continues to lag behind data acquisition capabilities. This slows data processing, collaboration, and quality control. Several large-scale database solutions [Reference Allan1] have been developed to manage imaging data, but the vast majority of laboratories rely on outdated methods of data transfer and management such as USB drives and personal computers. The success of high-performance cluster computing resources, when available, has yet to fully solve the data storage challenge. In this article, we compare common data storage solutions and discuss what can be implemented with different levels of financial and institutional resources, from cloud storage to institution-run storage servers.
Cutting-edge fluorescence and multiphoton microscopy provide a unique challenge for data storage and management. In our work we use two-photon light-sheet fluorescence microscopy (Figure 1), collecting whole zebrafish brain structure and activity data (Figure 2). An experiment can contain up to 50 axial slices, each covering a 400 × 400 μm area, collected with 3-second volumetric rate. This results in a 300–500 GB dataset per single sample. The data size is further increased when two or three spectral channels are imaged. Here we provide a description of the 250 TB storage server we have built and currently share between more than a dozen researchers. The system provides automatic backup and data access management. This guide can be used as a starting point for the transformation of data management practices toward more resilient and efficient data pipelines in laboratories that use modern microscopy tools. While our experiments are based on light sheet microscopy, the system described here can be used for any type of image data collection and storage.

Figure 1: Schematics of a light-sheet microscope. A two-photon light-sheet microscope consists of illumination and detection systems. Two-photon laser light (red) from a femtosecond source (such as a Ti:Sapphire laser) is scanned at a 1 kHz rate by galvo mirrors on to the illumination objective that forms a sheet of light. The fluorescence (green) in a model zebrafish sample is excited only within that sheet and detected using a high-NA objective on a sCMOS camera. Image courtesy of Thorlabs.
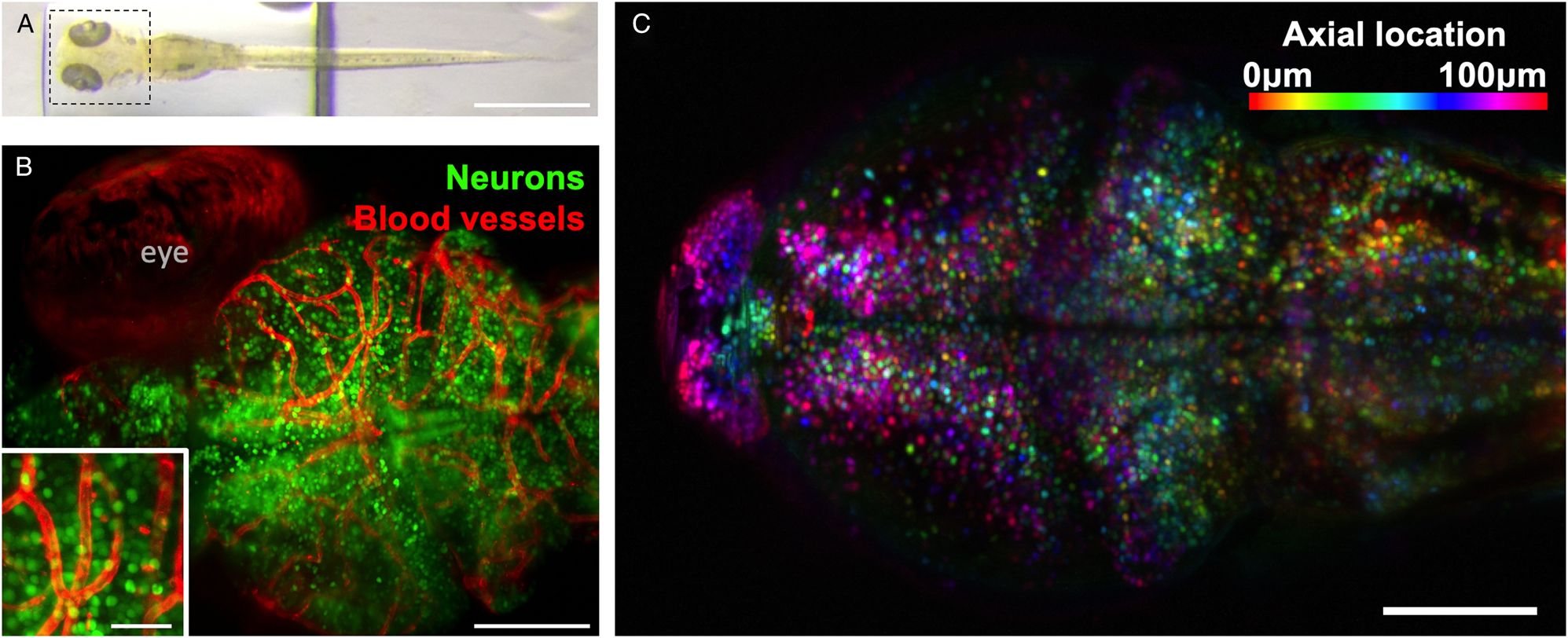
Figure 2: Example of zebrafish brain images collected using light-sheet microscopy. (A) Top view of a zebrafish larvae at 6 days post-fertilization. Scale bar 1 mm. (B) Maximum intensity projection of whole-brain imaging with diffraction-limited resolution of neurons (green) and blood vessels (red) allows longitudinal observation of vertebrate brain development. Scale bar 100 μm. Inset: magnified section of image, scale bar 25 μm. (C) Two-photon light-sheet imaging of fluorescent calcium indicator GCaMP6s allows recording activity of 50,000 neurons with single-cell resolution and 1-second temporal resolution. Color coding depicts neurons’ axial location within the sample. Images collected using Truong/Fraser setup at the University of Southern California.
Similar to other microscopy-oriented labs, we collect large amounts of data while simultaneously developing new experiments and data processing pipelines. Much of our work is in constant flux, and it is nearly impossible to reliably lock in a single data acquisition process for a long period of time. This flexibility requires rapid progress in development of our tools and puts additional pressure on the required data processing and storage infrastructure. The networks for data transfer from acquisition microscopes are often slow (less than 1 Gbps), and we rarely have centralized cost-efficient institutional storage capabilities. To use microscopy facilities more efficiently, we need networks that can transfer and store data at a speed of at least 1 Gbps, and centralized storage.
Current Practices
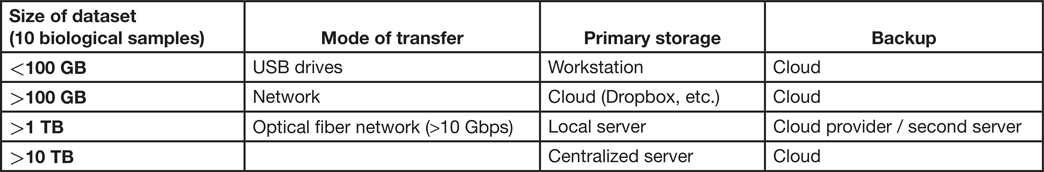
Today, research labs often use portable USB hard drives and cloud-based storage [Reference Tuyl and Michalek2]. These tools are poorly adapted for the data demands of modern microscopy. Here, we briefly review several solutions and provide recommendations on storage strategy depending on the size of the files requiring storage (Table 1).
Table 1: Recommendations based on scale. It is useful for labs to consider that there is more than one option to store data, and the strategy should be chosen based on needs. All different options can be used where appropriate. Here we provide a guide to selecting storage and backup strategy.
USB external hard drives.
Moving data using a USB hard drive or flash drive is very common in research. We can purchase storage very quickly and upload data immediately, since all research computers have USB ports. USB drives are very easy to use and allow for drag-and-drop operations for moving files from the data acquisition computer to a destination drive and/or data analysis workstations. Files may also be opened directly from the USB drive itself. Some devices on the market also provide increased reliability, such as portable external storage enclosures with multiple drives and duplication of files. An important advantage of USB drives is that they remove the necessity of network connectivity and allow “quarantine” of valuable research microscopes from the internet. The speed of data transfer can be relatively fast, ranging from approximately 1 Gbps to 5 Gbps with the newest flash-based storage and transfer protocols.
Disadvantages of USB hard drives include an increase in probability for data loss and difficulties related to data organization and sharing. A single USB hard drive may have a significant rate of failure (1–5% per year) or unrecoverable read errors (URE) (1 error per 12 TB of read data [Reference Hughes and Murray3]). Furthermore, stress on the connectors due to frequent usage may cause their failure within 1–2 years. USB drives can also serve as easy vectors for transfer of computer viruses, and drives should always be inspected prior to use.
As the number and size of data folders and files increase, the need for increased cataloging and organization of data across the various devices used for backup reduces the advantages of the “simple drag-and-drop” workflow. This is partially due to the hassle of determining which specific files may be located in which device, as well as the considerable time required for frequent transfers of large files. In addition, the bulky and wired nature of external hard drives further contributes to their inconvenience when having to use multiple drives. There is currently no easy way to streamline the process when using individual external hard drives for data backup. Data sharing is also difficult with USB external hard drives, as collaborators must have physical access to the drive or other means of sharing data.
In summary, USB hard drives are valuable when moving small volumes of data (approximately 100 Gb) where networks are not available, if a backup strategy is in place. As data volume increases the applicability of USB external drives decreases.
Cloud storage.
A number of “cloud” storage providers such as Dropbox, Google Drive, OneDrive, Amazon and others offer a simple workflow: a folder on a workstation is available to drag-and-drop data as an extension of a local file system, and data are automatically protected from loss. Theoretically, the storage volume is unlimited, or specific recommendations may be given depending on usage and/or price. Institutions often offer free access to cloud storage solutions, which is appealing to research groups. Sharing is usually built in as an easy “share by link” function, and, since the service is Internet-based data, it can be shared virtually any place in the world. Data backup and version control (rollback) are great features that provide the ability to reverse any accidental deletion or overwriting of valuable files.
Unfortunately, limitations on cloud storage often occur when the amount of data being stored reaches beyond a few terabytes. Data transfer also depends on network stability and speed, which are often inconsistent. This inconsistency in data transfer, along with the common case of sharing a single acquisition computer for collection of data in a research setting, further limit the functionality of cloud storage, which requires uploading from a workstation. Furthermore, collaborative storage, usually a necessity among lab members, is often difficult to set up. Although storage is often advertised as unlimited, it is rarely truly unlimited. Cloud storage providers often “throttle” or decrease bandwidth after a certain amount of data has been moved in a given period of time (for example, 15 GB per month). We faced this issue when “unlimited” cloud storage decreased the speed of an upload by a factor of 10 after the first 100 GB of data were uploaded. Reduced transfer rates may completely halt the use of cloud storage if not planned in advance or when working with large datasets (>100 GBs). Some providers may also offer free institutional access but limit data storage for 24 hrs. Even with unlimited cloud bandwidth, the speed of data transfer can still be limited by fixed institutional internet speed, which is often below 1 Gbps. In this situation, just transfer of a single 10 Gb file may take 30 minutes.
In summary, cloud storage may be appropriate when data size for a project is limited to a few GBs and the total amount of data per project is below 1 TB, data acquisition computers are attached to the Internet, and only a few people work with the datasets.
Dedicated servers for network-attached storage.
Network-attached storage (NAS) is a dedicated data storage computer, or bank of computers, located on the same network as the acquisition microscope and image processing workstation. To the user, the storage is presented as a network drive allowing simple “drag-and-drop” workflow as a local drive or USB hard drive functions. A major benefit is that NAS physical storage is independent of data acquisition and image processing systems. Having a dedicated storage server allows gradual storage expansion as required without rebuilding the system from an initial ~100 TB up to and above 500 TB. When the NAS, acquisition microscope, and image processing workstation are physically close to each other, affordable fast networking with speeds of 10 Gbps to 100 Gbps can be installed minimizing delays in data transfer.
One approach is to place a data processing server in the same computer rack with the storage server connected by a 10 Gbps network, which can be accomplished for less than $1,000 (Table 1). This allows the end user to access the data processing machine remotely. Backup can be implemented using a second server or via the internet to cloud storage as described above. The process of data transfer to the cloud can be automated and hidden from the end user. The administrative interfaces available for such systems make it a relatively simple task for a non-professional to manage data storage and transfer, and the stability of the system ensures that little service is needed after initial installation. The NAS solution can be securely shared between several groups, decreasing costs for the individual research laboratories as described below.
It is possible for individual laboratories or small groups of investigators to develop and share NAS capabilities with a relatively low investment of time and money (Table 2). Management is possible by the laboratory members, but in a better scenario, laboratories should purchase a dedicated server and manage the system with assistance from the IT department, since fast networking between floors or buildings will most likely require institutional support. Access from outside the institution can be restricted by the IT department firewall in order to limit vulnerability.
Table 2: Estimated cost of server for storage of 250 TB. Building dedicated storage can be expensive, and we provide an example of budget using commodity parts. Pricesare in USD.
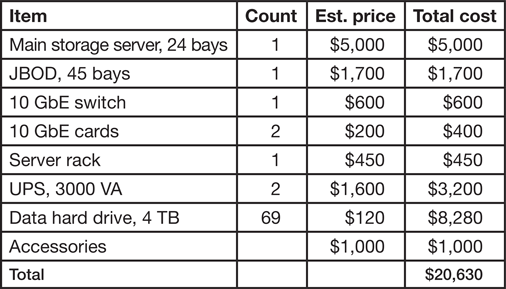
In summary, the original price of a NAS system ($20,000 for 250 TB, Table 2) can be intimidating, and institutional IT assistance for setup may be required. Given these limitations, acquiring a personal laboratory NAS is advised only when individual datasets are larger than tens of GBs, the total amount of data storage is larger than 10 TB, or when storage needs to be secure, fast, and expandable. Details of one implementation of a NAS system is provided in the next section.
Implementation of a Resilient 250-Terabyte Storage NAS
To provide storage of large volumes of imaging data, we have built a centralized storage server using commercially available hardware and free open-source software. The original goal was to provide storage for fluorescence light-sheet microscopy data and specifically whole-brain imaging of zebrafish neural activity. These experiments routinely generate datasets of approximately 500 GBs. Since implementation three years ago, the server has turned into a shared resource and now serves more than a dozen projects and researchers without data loss or service interruption. To provide this resiliency, we had to account for potential hardware failures, power loss events, and random errors during data reading and writing.
Protection against single-drive failure.
To create large centralized storage, we pooled together up to 70 bare spinning-disk hard drives (internal SATA drives) for a total storage capacity of 250 TB. When working with a large number of high-capacity drives, we faced the challenge of random errors that can corrupt stored data [Reference Schroeder and Gibson4]. Use of redundant array of independent drives (RAID) is a common strategy to prevent data corruption. Instead of a dedicated RAID card, we used a software solution relying on a file and operating system (OS) that are optimized to consolidate a large number of drives, monitor the health of volumes and data, and regularly scrub data to correct any errors. For this purpose we picked the freely available open-source FreeNAS [Reference Sims5] system, which comes with all features of the mature server-grade FreeBSD OS and ZFS filesystem and adds a user-friendly web interface for configuration and management of the storage.
Physical design and power redundancy.
We used server-grade hardware to implement storage. The main server has 24 drive bays with each drive bay accepting standard 3.5-inch hard drives of 4 TB capacity each. The server is equipped with two Xeon processors and 256 GBs of RAM. We used an expansion box with 45 bays, colloquially referred to as Just a Bunch of Disks (JBOD), where all drives are attached to the host-bus adapter (HBA). The JBOD is connected to the main server using a single SAS3 cable. Data drives are organized in RAID6 stripes of six drives. The system drives are two 120 GB solid-state disks (SSD) in mirror configuration (RAID1).
Network connectivity was established using 10 GbE cards that can accommodate copper wire or optical fiber connectors. Recently, servers have started to provide built-in 10 GbE connectors on the motherboard. The storage server is connected to two networks: a local network between the data storage system and a light-sheet microscope using a 10 Gbps transfer, and a general-use 1 GbE connection to the rest of the university network. Thisdual connection ensures that even if the university network is unavailable due to failure or maintenance, we can still use the storage server to store large data sets from the light-sheet microscope. The local 10 GbE network was set up using an affordable 8-port switch, and it connects in one rack the storage server, acquisition computer, and analysis workstation.
The main data server, JBOD, and other computers mentioned have redundant power supplies, which allows us to use two uninterruptible power supply (UPS) units running at half-capacity each. If one UPS fails, the second will be able to provide power to the whole setup. When picking the UPS power rating we estimated a 7-minute run on the battery, which is the time interval for the power generator in the building to be activated. Installing this power required contract work with the institution. The final assembly of servers and UPS is depicted in Figure 3.

Figure 3: Organization of servers. From the bottom up: (1) Two redundant UPS units, (2) main storage server, (3) JBOD expansion, (4) analysis computer, (5)web server for https://fliptrap.org. All servers are mounted in a standard-sized 19-inch-wide server rack using supplied rails.
Data snapshots and backup.
When working with data we may accidentally delete or overwrite files. To partially protect against these actions, the ZFS file system can take “snapshots” of data, recording changes made since the last snapshot. Any file in the ZFS file system can be “rolled back” to the previously available snapshot instantaneously. We used the FreeNAS system's automatic scheduler to set up daily snapshots that are kept for a month. The data backup is implemented as a daily transfer of snapshots to another server that is also running the ZFS operating system, that is, 6 TB drives organized as 8-drive RAID6 stripes in a 36-drive box, 2x Xeon 2.2 GHz CPU, 64 GB ECC RAM. The backup can be also automatically stored using one of the cloud providers.
Conclusion
Current data storage methods lag far behind the development of modern microscopy tools. It makes processing of data difficult and data sharing slow, while the amount of imaging data continues to increase. Using USB hard drives, cloud storage providers, or shared network-attached storage servers should be carefully designed, and benefits and drawbacks should be considered. Here we provided our implementation of a shared expandable storage system as a starting point and an approximation of such a system's cost. Ideally, the laboratories that work with significant amounts of data will consult and receive help from their institution, potentially creating a shared-pooled resource as economy of scale makes services cheaper.
Acknowledgements
We would like to thank Tom Limoncelli, Dr. Francesco Cutrale, Dr. Jacqueline Campbell, Dr. Sandra Gesing, and Tom Morrell for valuable discussions.








