Introduction
Historically, advances in imaging have spawned significant trends in biomedical research. Invention of the compound microscope by Hans and Zaccharias Jansen in the late 16th century enabled visualization of objects and events beyond the resolution of normal human observation. Likewise, discovery of X-rays by Wilhelm Roentgen in 1895 enabled visualization of occult internal structures and processes obscured by tissue opacity and led to the field of medical imaging that uses non-invasive imaging of internal structures at a macroscopic level [Reference Mankoff1]. More recently, development of fluorescent dyes and molecular targeted imaging have led to the development of a multitude of new methodologies for the observation of physiological and pathological events in systems biology. Despite the innumerable contributions of these imaging technologies in the advancement of biomedical research, there remain several translational questions that fall between microscopic and macroscopic approaches. The need for technology to bridge this gap for both morphological and functional imaging has been a key goal for some time, especially in the post-genomic era where vast libraries of new molecular entities (NMEs) as candidates for novel therapeutics have been developed. However, despite the escalation in screening methodologies, the true performance of these compounds in intact biological systems remains challenging [Reference Szymański2]. The role of high throughput and high content screening in drug development is evidenced in FDA guidelines and review papers [Reference Zock3]. Indeed, the more advanced models (such as whole zebrafish) are used for high-throughput assays in a whole-body context. However, these model organisms are far from the biological complexity found in mammals, especially humans.
Confocal laser endomicroscopy (CLE) is a set of imaging modalities that combines endoscopy and confocal microscopy to bridge the gap between macro and micro imaging modalities [Reference Robles-Medranda4–Reference Moussata7]. In this article, we describe fluorescence in vivo endomicroscopy (FIVE), a cutting-edge CLE that enables non-destructive and temporal visualization of microscopic cellular architecture and cellular behavior in living organisms. In a follow-up article we will describe several applications of CLE and FIVE.
Development of Confocal Endomicroscopy
In the 1950s, Marvin Minsky invented and patented the confocal microscope. His goal was to map connections within neural networks, 3-D visualization of which was very difficult using microscopy techniques of the day. The invention was based on aligned pinholes for illumination of, and detection of light emanating from, a single focal point. This optical geometry, with the illumination and detection paths sharing a conjugate focus (hence the term “confocal”), effectively constrains the detector from “seeing” anything but that single focal point. This can be thought of as producing a single pixel in 3-D space. Forming an image requires this point to be scanned across the sample (typically in a raster pattern), essentially serializing an image comprised only of in-focus points. Thus, it is a sectional image, and if the focal plane is shifted in depth, 2-D images at various planes can be used to construct a 3-D array of pixels to generate a 3-D data set.
Minsky faced many constraints in condensing the idea to practice. Digital image acquisition, sensitive light detectors, lasers, and raster scanning mirror actuators were all yet to become available. Nonetheless, using various means to scan the light path or sample itself, he was able to achieve proof of principle in several different forms. The apparatus was fiddly to set up and not practical for general utility among microscopists. However, the principle was established and is the foundation for all confocal microscopes today. Six decades later, advances in computing and lasers have made commercial confocal laser scanning microscopes (CLSM) a reality, and imaging of a tissue in 3-D is commonplace. These CLSM devices are large, mounted to rigid optical benches, and mandate complex optical alignments. The subject must be stabilized on a microscope stage and therefore in vivo imaging applications are limited.
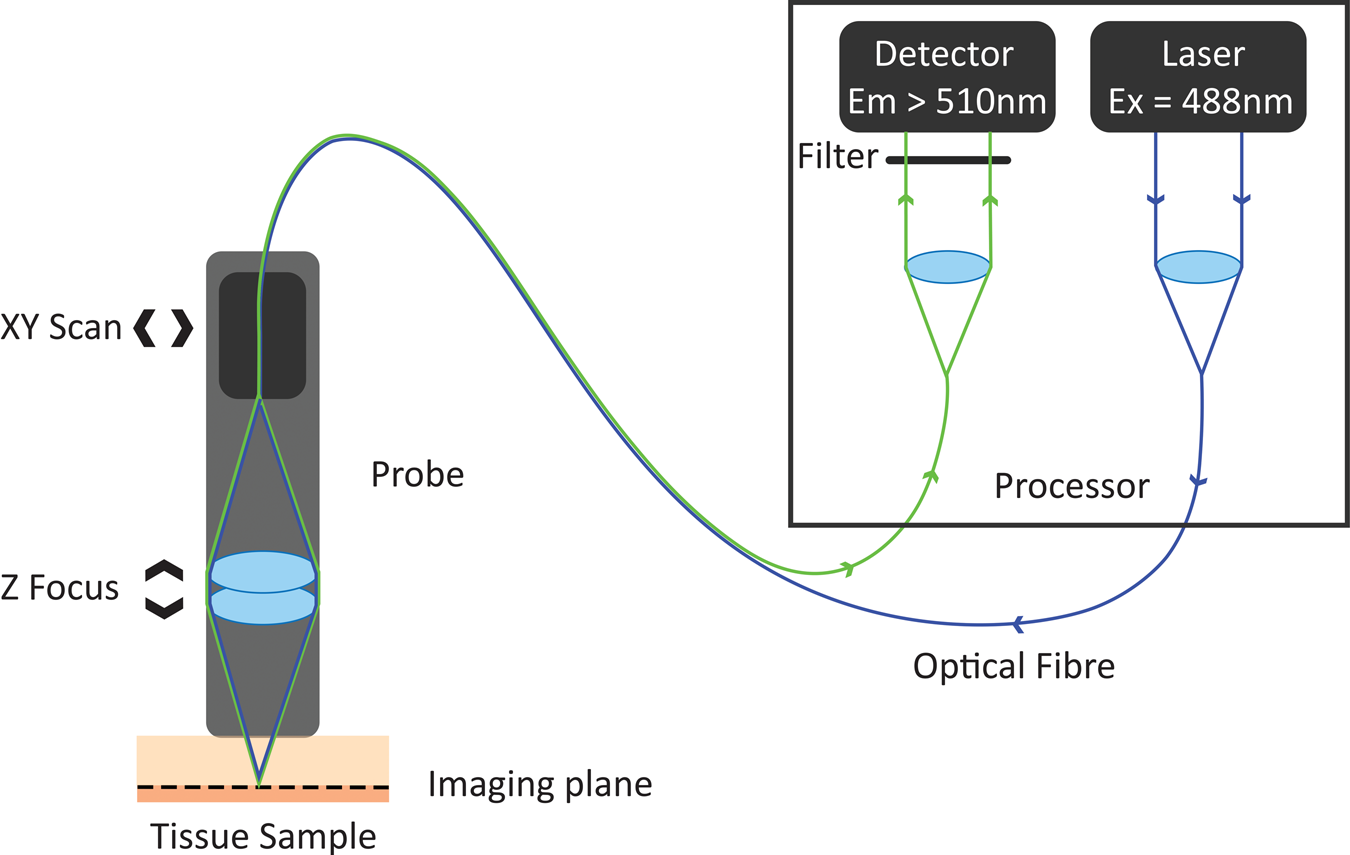
In the 1980s, Australian inventor Martin Harris realized that the tip of an optical fiber with certain characteristics could simultaneously form the illumination and detection pinholes of a confocal microscope. As the illumination and detection apertures are the same physical entity, no complex co-alignment of pinholes to illumination paths was required, simplifying the optical design. Harris and his colleague Peter Delaney recognized that this invention provided pathways for the miniaturization of confocal microscopes and led to the invention of confocal laser endomicroscopy (CLE) [Reference Delaney8]. These initial developments were based on two different scanning technologies. One approach was to sequentially scan individual optical fibers in the proximal end of an optical fiber imaging bundle, with distal tip optics focusing the light into the sample. This approach gave rise to fiber bundle-based CLE. The other approach was to physically scan the tip of a single optical fiber behind a lens system that relayed the path of the optical fiber to a conjugate focal plane in the tissue. The scanning single-fiber technology was named fluorescence in vivo endomicroscopy, and the short form FIVE was coined (Figure 1).
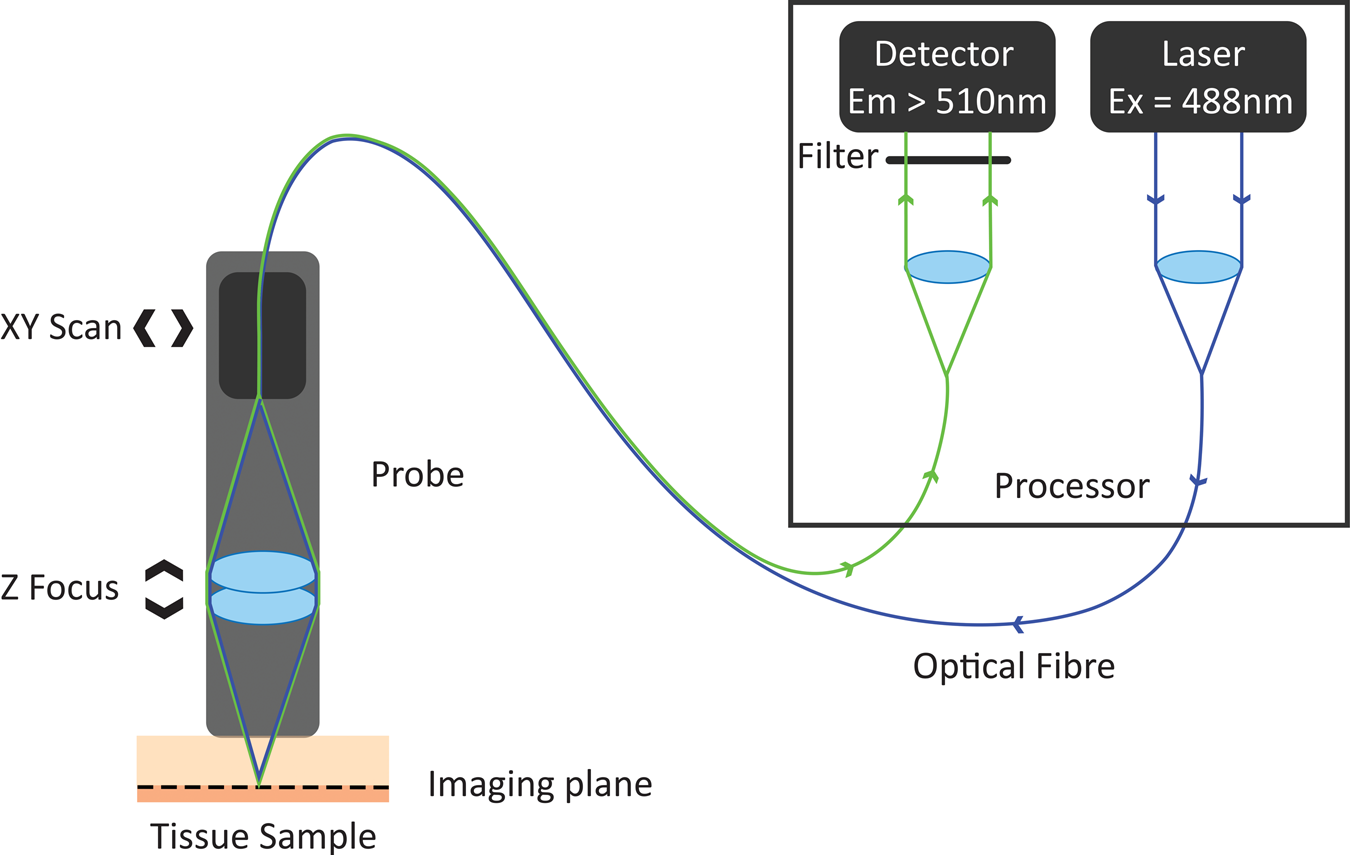
Figure 1: Working principle of FIVE. Light from the laser is focused into an optical fiber. The tip of the fiber is moved in a raster pattern, scanning light across the back of a projection lens. The lens focuses light to a spot on the sample. Some of the fluorescent light from that spot is captured by the lens and focused into the optical fiber tip. The fluorescent light is split from the optical fiber and measured by a detector. A computer assembles the image based on the detected light intensity at each 3-D focal point traversed during the scan.
Fiber Bundle Endomicroscopy
Sequential scanning of individual fibers in coherently fused fiber optic bundles is an attractive idea, as the miniaturized microscope tip has no moving parts, and their diameters can be as small as the fiber bundle. However, this approach limits resolution, and higher resolution requires more signal and more fibers, which increases the tip diameter significantly. Moreover, a bundle fiber imposes a fundamental limit on image resolution as there can be only one focal point per fiber. Field-of-view can be varied by varying the magnification of the projection lens. Although different devices can therefore feature different magnification, the information content of the image is still constrained by the number of pixels. Using a bundle with more pixels can yield higher information content, however the size of the individual fibers is dictated by the light guiding requirements for each fiber to act as a confocal pinhole. This means that the size of the device scales with the number of fibers. Bundles having more than 30,000 elements become quite stiff and no longer suitable for endoscopy. In reality, most bundle endomicroscope probes are 30,000 pixels or less, and these can be spread over a very small field-of-view to extract cellular detail or spread across a usable field-of-view at the expense of resolving individual cells. A further compromise in bundle-based probes is that light only travels in the core of each fiber, so there is information missing from between the fiber cores. Manipulation of the raw image data is therefore required to “smooth” or de-pixelate the image. Hence, while probe endomicroscopy based on fiber bundles offers access and convenience to deploy via working channels of common endoscopes, it comes at the expense of performance of field-of-view and cellular resolution. This is evidenced in outcomes of clinical trials, which have failed to achieve the requisite American Society for Gastrointestinal Endoscopy (ASGE) efficacy threshold for Preservation and Incorporation of Valuable Endoscopic Innovations for adoption into clinical practice in some applications [Reference Thosani9].
Scanning Single Fiber Endomicroscopy/FIVE
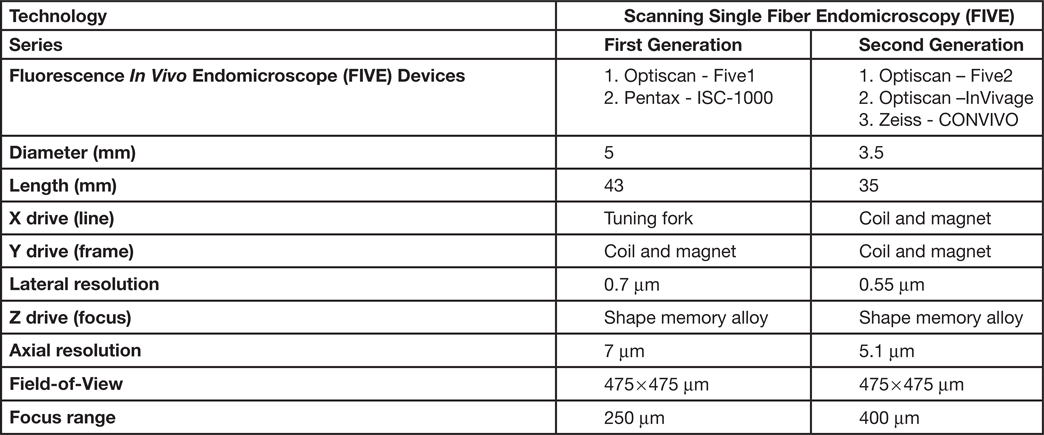
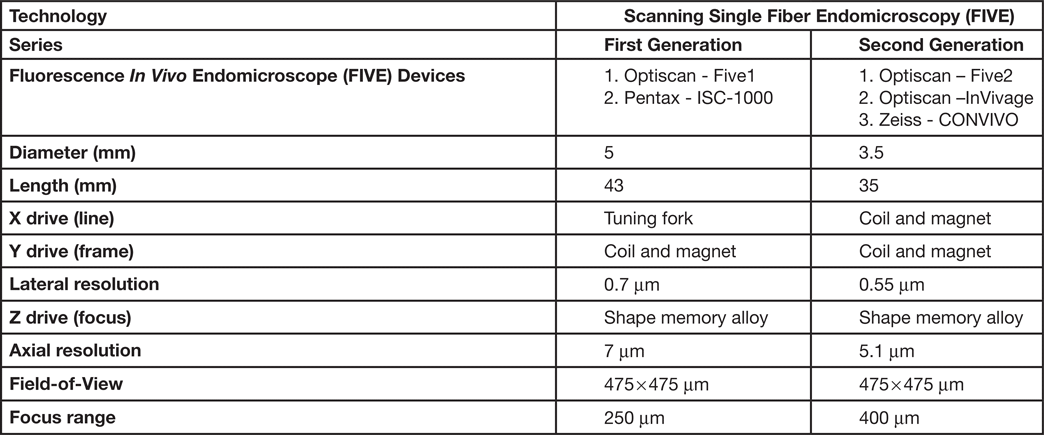
The scanned fiber approach to miniaturizing confocal microscopes overcomes the sampling and optical resolution limitations of bundle fiber endomicroscopy, as sampling rate can be matched to the optical resolution of the lens system and the image field-of-view. This enables collection of megapixel images at submicron lateral resolution with a large field-of-view. Additionally, this approach provides the ability to move the focal plane in the Z-axis. This is important as cell layers within tissue can change dramatically over a few microns (Figure 2). The ability to perform optical sectioning of tissue enables image capture from each cell layer, producing a layer-by-layer 3-D image stack of the tissue (Figure 3). The advantage of performing optical sectioning with submicron resolution gives an edge to FIVE over other CLE technologies, as one can visualize the histoarchitecture in X, Y, and Z planes, offering a better understanding of tissue architecture and the disease. However, these advantages come at a cost of added complexity of the mechanism in the tip of the endomicroscope. Several commercial devices with scanned fiber mechanisms have been produced. Their specifications and performance are summarized in Table 1.
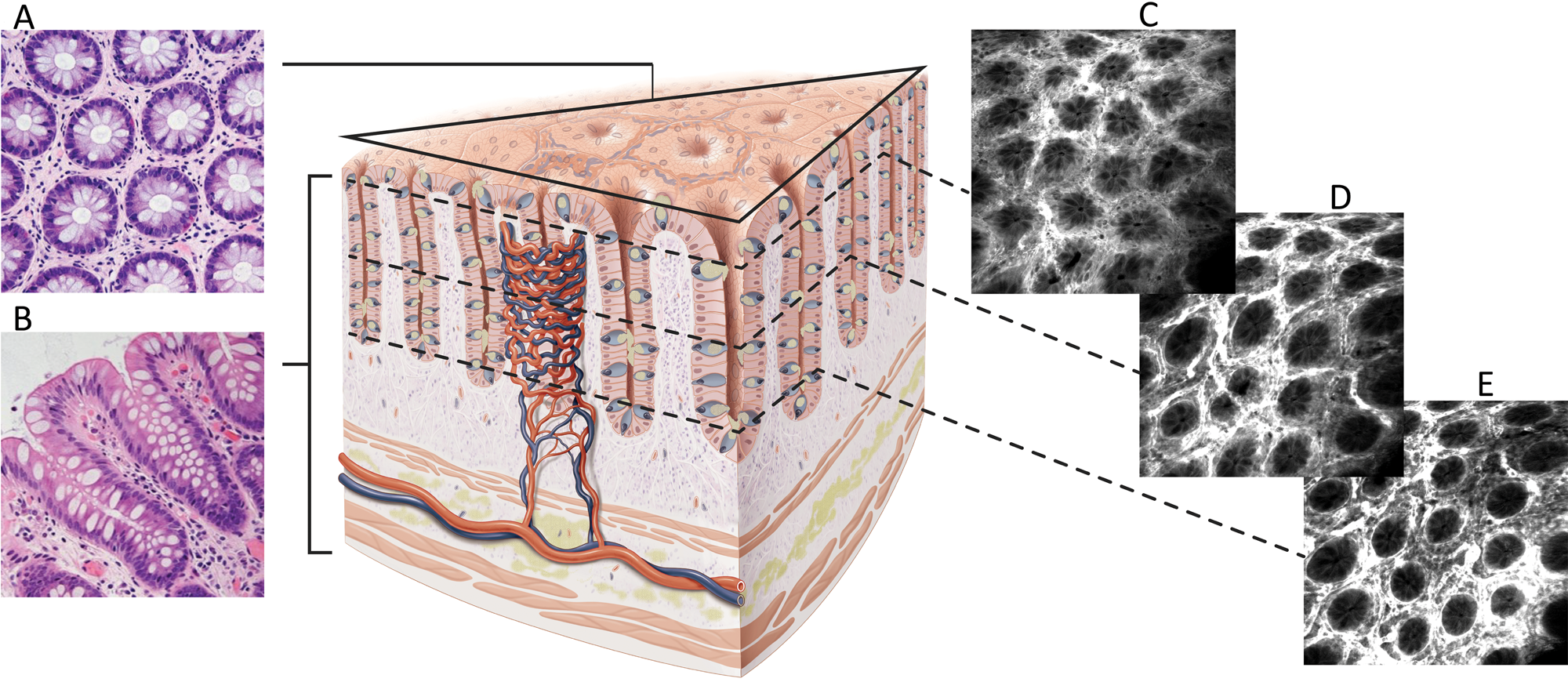
Figure 2: Difference between conventional hematoxylin and eosin (H&E) histology and CLE images. A. En face histology representation to the same as CLE image orientation. B. Conventional H&E transverse section of fixed and stained tissue. C–E. FIVE optical sections of living gut showing images at different depths from the mucosal surface.
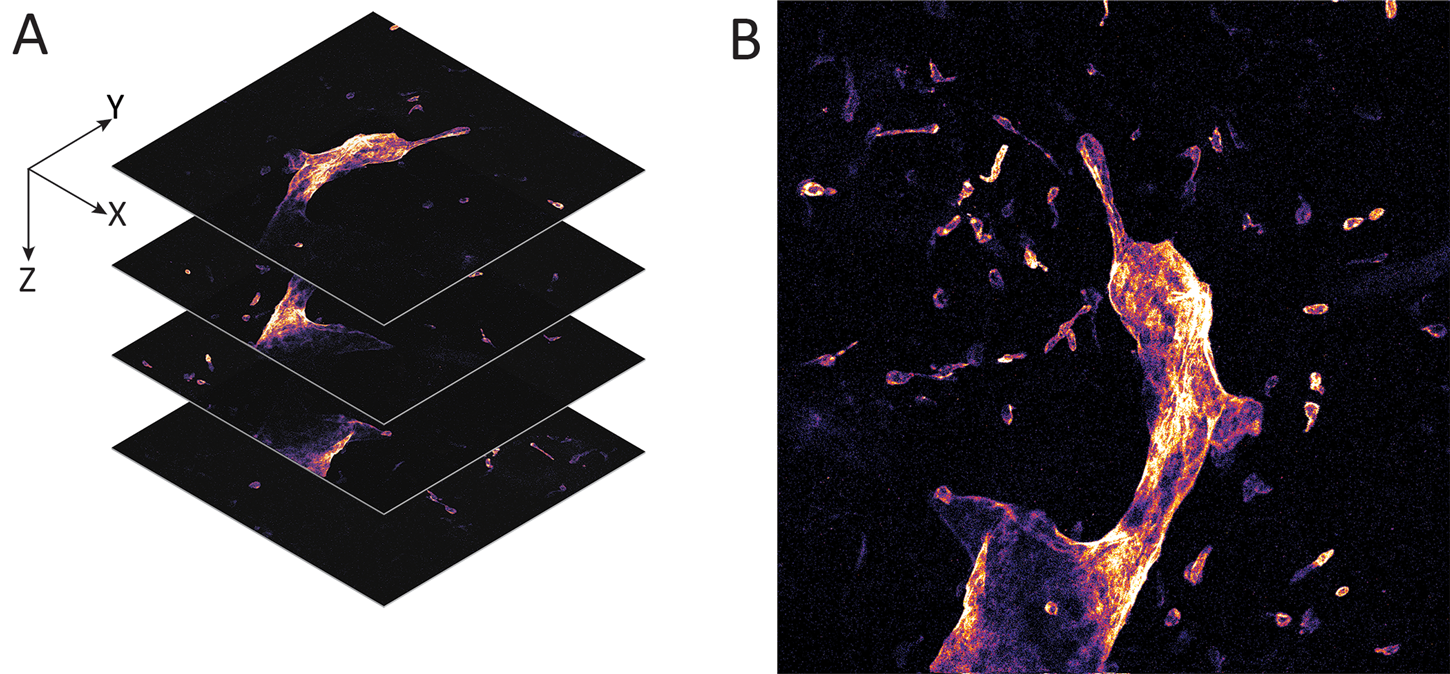
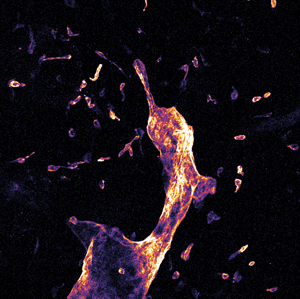
Figure 3: Whole mount visceral adipose tissue stained with LYVE-1 Alexa Fluor 488. A. Schematic representation of a 3-D stack collection by confocal endomicroscope. B. Maximum brightness projection of the collected Z-stack. Published with permission of Enyuan Cao, Monash Institute of Pharmaceutical Sciences, Monash University.
Table 1: Types of FIVE systems used in clinical and preclinical settings
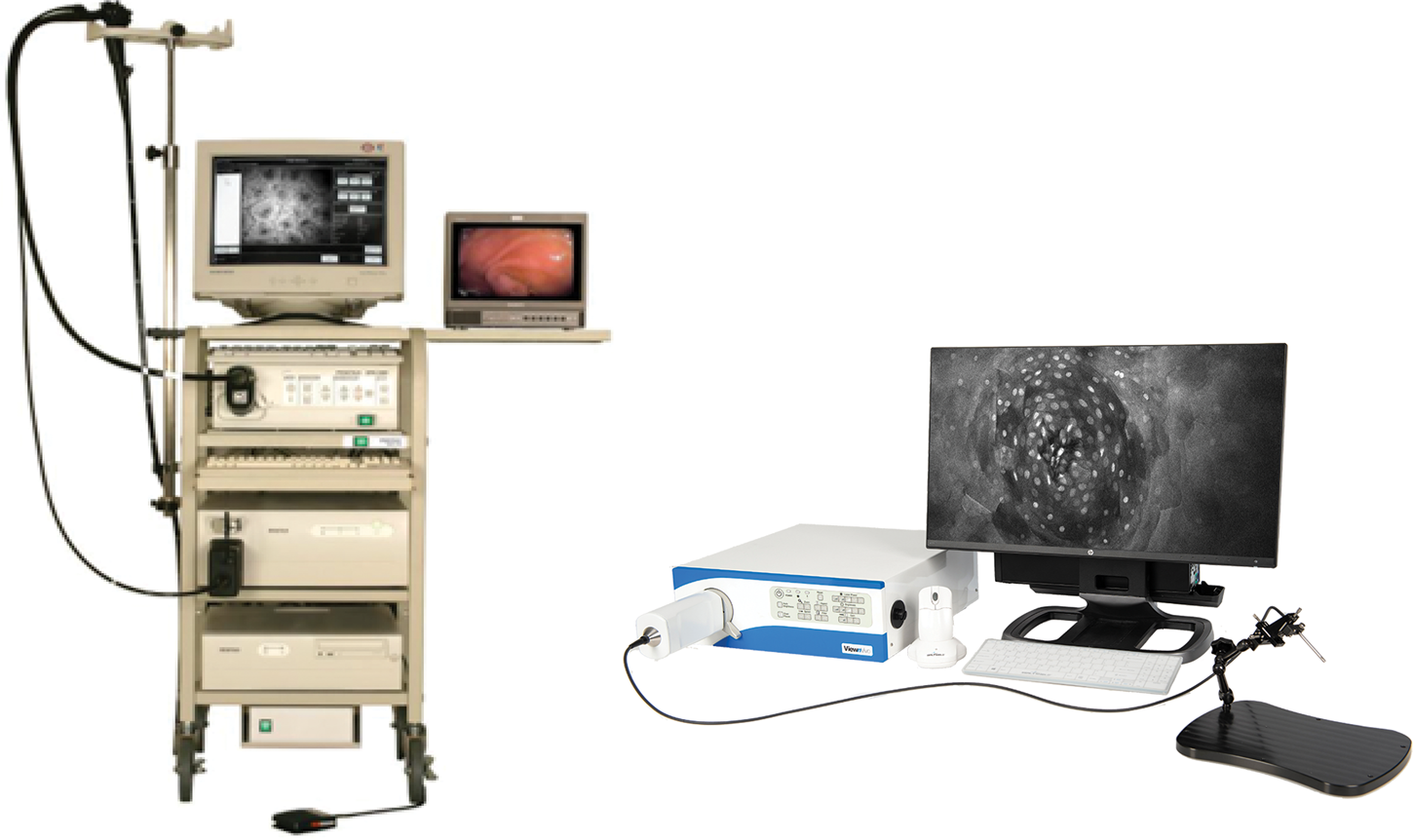
Because of the advantages of FIVE over other CLEs, its principles were adopted in the Fluorescence In Vivo Endomicroscopy (FIVE) instrument series. The first generation of the FIVE instrument series was integrated into the Pentax ISC-1000 endoscope (Figure 4A). It was a highly miniaturized scanner with an integral electronic focus mechanism enabling the field of gastrointestinal endomicroscopy. A handheld rigid version of this device was also used as a stand-alone laboratory instrument, named FIVE1, with numerous preclinical and veterinary applications. The second generation of FIVE instruments was triggered by the requirement for a smaller, more robust device with higher resolution, faster frame rates, and more precise and explicit Z-depth (focal plane) control. In the second-generation devices, line and frame scans were combined in a single mechanism (Figure 4B). The improved Z-depth control was based on the same technology as the first-generation devices but refined and configured with improved positional control. Optimization of the optics improved the resolution with a 30% reduction in diameter, 70% reduction in overall volume, and double the working distance. This scanner is used in Optiscan's FIVE2 laboratory endomicrosopes, Optiscan's InVivage™ dental imaging device, and Carl Zeiss Meditec's CONVIVO™ for use in neurosurgery.
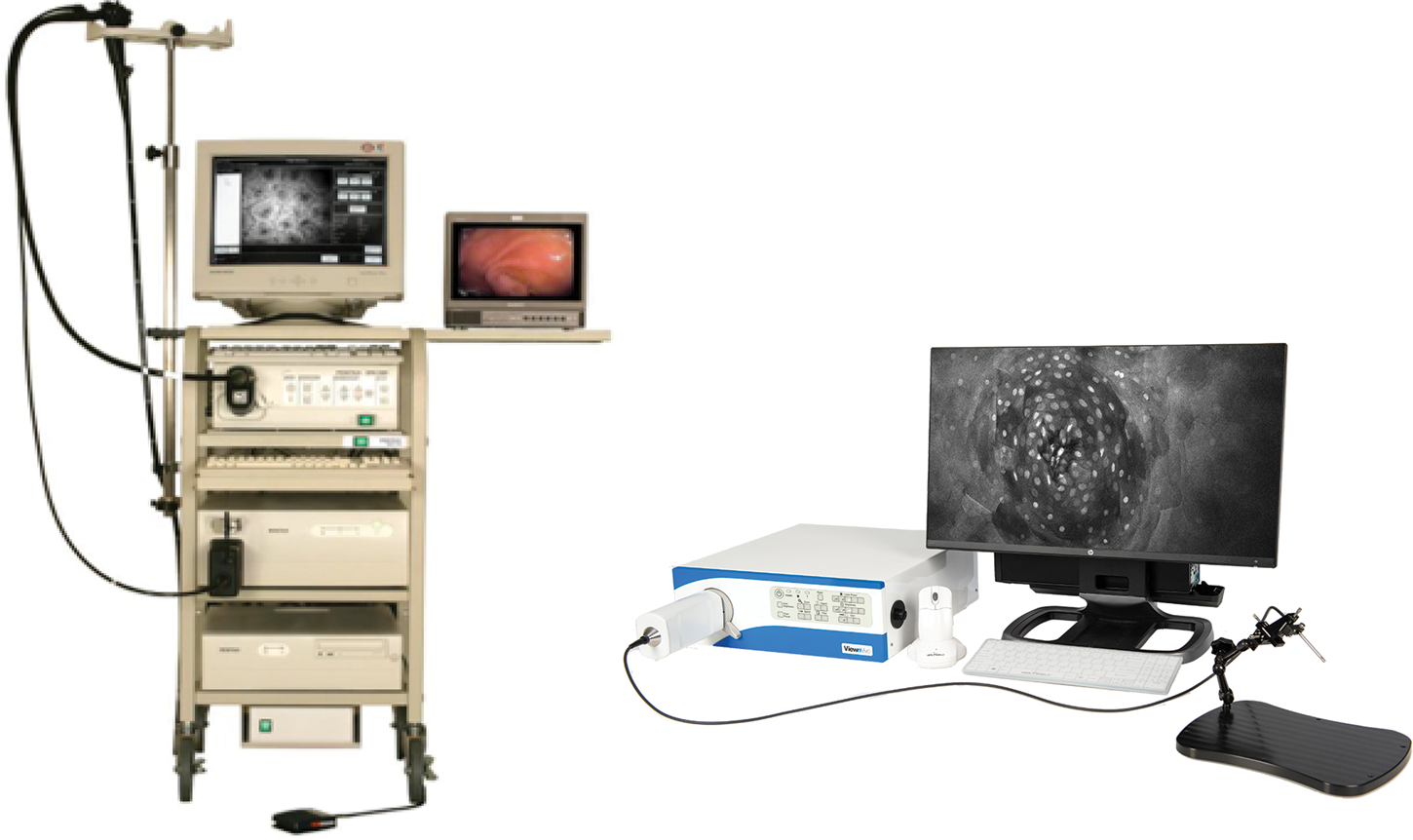
Figure 4: First- and second-generation devices from Optiscan. Left. Pentax ISC-1000 integrated with Optiscan's confocal scanner. Right. Optiscan's second-generation laboratory device, FIVE2 (ViewnVivo).
FIVE versus Pathology
An important question in the development of FIVE was the degree of utility of the images obtained. FIVE images are not the same as a photograph of a microscope slide. They are an intensity map measuring fluorophore concentration variations in living tissue, and this is dependent on the characteristics of the fluorophore, for example, how it is applied, what are the pharmacokinetics within the tissue being imaged, and the distance between the image plane and the probe. The questions about the usefulness of FIVE images can be addressed in three parts. Firstly, what is the correlation between FIVE images and classical histological images? Secondly, would medical practitioners need pathologist-equivalent expertise to understand the FIVE images, and thirdly, what do the images tell us about microscopic processes in the living tissue being imaged, including observations that cannot be made ex vivo.
FIVE images are typically en face and parallel to the surface of the tissue being examined (Figure 2). The field-of-view is nearly 0.5 × 0.5mm2, so the patterns of cells at the tissue surface can be compared over a microscopically large area. This compares favorably with the typical histology slide thickness of about 5 µm. To explore the usability of FIVE images, a study looking at cervical intraepithelial neoplasia (CIN) showed that characteristic tissue architectural patterns enabled even non-experts to easily recognize image features consistent with high-grade CIN. Features examined included increased nuclear density and hyperchromatic and pleomorphic cell nuclei, providing a high degree of accuracy in identification of dysplastic tissue [Reference Tan10]. In another example, physicians without any FIVE experience were trained using a set of FIVE video clips. Upon completion of the training, their accuracy for identification of intestinal metaplasia in FIVE images was 80%, with substantial interobserver agreement [Reference Pittayanon11]. In yet another learning curve study, images obtained with FIVE from gastrectomy specimens were assessed by trained endomicroscopists, and the sensitivity and specificity of assessment of intestinal metaplasia was 95.2% and 93.3%, respectively. This was significantly higher than for inexperienced examiners (61.9% and 62.2%, respectively) [Reference Lim12], indicating that there is a learning effect for interpretation of FIVE images, but that even inexperienced examiners were able to identify disease in nearly 2/3 of images.
Limitations of FIVE
There are limitations to what can be imaged using any in vivo microscopy technique. They are due to a combination of the optical properties of tissues, the ability to physically access tissues of interest, and the abilities of fluorophores to label and contrast features of interest. As with any confocal microscope, the imaging depth of FIVE is limited by the optical characteristics of the tissue, such as refraction, scattering, absorption, and opacity. Connective tissues present fine-meshed refractive index boundaries, and absorption of blue light can reduce effective imaging depth to between 50–200 µm in most tissues. Some tissues, such as brain, are highly scattering due to the close-packed lipid-aqueous boundaries associated with neurons and their processes, so usable images are usually only obtained to a depth of just a few tens of microns, and the best images are obtained close to the tissue surface. In addition to scattering, quenching also reduces image quality as imaging depth increases. This is where the fluorescent dye used to provide contrast, or other molecules, absorb some of the fluorescence emitted. This is particularly problematic when there is a concentrated pool of fluorophore at the tissue surface. Autofluorescence is another factor to consider when imaging in vivo. This can be minimized by keeping laser power low and using optical filters to collect detected light from the peak emission band of the contrast agent used. Optimal imaging parameters must balance fluorophore concentration, laser power, and detected wavelengths.
Physical access to tissues of interest is another limitation, with mucosal epithelium being the most amenable for FIVE. Pentax ISC-1000 endomicroscopes can image most of the upper and lower GI tract. Rigid probes can access some tissues of the mouth and reproductive tract through natural orifices. Rigid devices can also be used to image surgically exposed tissues or in keyhole surgery. However, it is generally only tissue and organ surfaces and surgical wound beds that can be accessed. Developing smaller devices and integrating them into specialized devices increases the number of tissues that can be imaged and provides the potential to better image tissues like airways, bladders, and thoracic or peritoneal cavities. An additional challenge for FIVE is the limited number of non-toxic clinically approved fluorophores. While intravenous fluorescein provides good images in many contexts, its lack of specificity reduces its usefulness. The development of fluorophore-tagged molecular markers is a potential game-changer, as they provide the ability to image specific targets in vivo. An example of this is PARPi-FL, a cancer detection biomarker developed at Memorial Sloan Kettering Cancer Center in New York [Reference Kossatz13–Reference Kossatz14].
Summary
FIVE instruments are potentially valuable tools for the in vivo imaging of cells, tissues, and organs. It has enabled the visualization of dynamic processes in vivo including apoptosis, cell turnover, mucosal barrier functions, and interaction of the microbiome with the mucosa. It has also been used for looking at drug delivery and the binding of specific molecular markers in vivo. In part 2 of this series, we will discuss the applications and the future of the FIVE technology.