Introduction
Cholera is a severe form of waterborne acute dehydrating diarrhoeal disease which is well known for its epidemic and pandemic potentials [Reference Kaper, Morris and Levine1, Reference Mukhopadhyay, Takeda and Nair2]. Cholera continues to be a major public health problem, particularly in developing countries where access to potable drinking water and hygienic sanitation [Reference Harris3, Reference Ali4] remains inadequate. Toxigenic strains of Vibrio cholerae serogroups O1 and O139 are the causative agents of cholera, and exhibited several changing patterns in the biotype as well as in drug resistance for better survival and infection ability [Reference Kelly-Hope5, Reference Raychoudhuri6]. In 2014, 42 countries reported to the World Health Organization (WHO) a total of 190 549 cholera cases with 2231 deaths, resulting in an overall case fatality rate (CFR) of 1.17%. About 55% of all reported cases originated from Africa followed by Asia. In India, 4031 cholera cases with 21 deaths (CFR, 0.52%) were reported from 12 States and most of these cases (49%) were reported from West Bengal [7].
Cholera exists as a seasonal disease in India especially in Kolkata, the capital city of the state of West Bengal located in the Gangetic delta and has been greeted as the ‘homeland of cholera’ [Reference Sengupta8–Reference Sur10]. Cholera cases occur round the year in this city with different magnitudes. This disease becomes more pronounced in those areas where there is post-monsoon water logging or overflow of drains contaminating surface water bodies due to inadequate drainage system [Reference Sur11].
During July 2015, there was a heavy rainfall in Kolkata which led to overflow of many drains and water logging at certain parts of this city. In the first week of August, Infectious Diseases and Beliaghata General (ID&BG) Hospital of eastern Kolkata witnessed a huge increase in the number of diarrhoea patients requiring admission for treatment. The National Institute of Cholera and Enteric Diseases (NICED), Kolkata, has been conducting diarrhoeal disease surveillance in this hospital for almost past two decades, where systematically every fifth hospitalised diarrhoea case on two selected days in each week is recruited [Reference Nair12]. In response to this flood situation, scientists at NICED analysed surveillance data of a subset of the diarrhoea patients admitted to ID&BG Hospital from Kolkata Municipal Corporation area during July–August 2015. The investigation was undertaken to confirm existence of any diarrhoea disease outbreak, to recommend appropriate preventive and control measures, to identify the aetiological agents of outbreak, to determine the antimicrobial susceptibility of the isolated organism against a range of antimicrobial agents and to determine the genetic relatedness of the isolated strains during this period.
Materials and methods
Epidemiological analysis to understand the existence of a diarrhoeal outbreak
Information on selected demographic, socio-economic and clinical profiles was collected by trained NICED staff using the structured questionnaire from the patients or the parent caregivers in case of children through the routine surveillance of NICED. The clinical assessment was made according to WHO guidelines [13]. Data were checked manually and subsequently entered in the surveillance-specific database designed at NICED. Analyses were performed using the statistical software STATA SE ver. 8.2 (Stata Corp, Texas, USA).
The mean of hospital admissions from Kolkata metropolis area during first 2 weeks of August of 2007–2014 were also compared to understand the magnitude of the situation in 2015.
Laboratory investigation
Stool specimens from the patients were collected using rectal catheters and were transported within 2 h of collection to the laboratory at NICED for further processing and testing as described below.
Stool culture
The samples were processed for isolation of common enteric pathogens such as Vibrio spp., Salmonella spp., Shigella spp., diarrhoeagenic Escherichia coli and Campylobacter spp. [14]. Alkaline peptone water was used for the enrichment of V. cholerae, whereas Gram-negative broth (Difco, BD, USA) was used for the enrichment of Salmonella spp. and Shigella spp. Enrichments were sub-cultured on thiosulfate-citrate-bile salts-sucrose (TCBS) (Difco) for V. cholerae and Hektoen enteric agar (HEA) and xylose lysine deoxycholate agar (XLD agar) (Difco) for Salmonella spp. and Shigella spp. All plates were incubated overnight at 37 °C. The yellow-coloured colonies on TCBS media for suspected V. cholerae were subjected to standard biochemical tests, including sugar fermentation in triple sugar iron agar, and oxidase production. Detected V. cholerae were identified to belong to O1 serogroup through slide agglutination test using antisera kit (Difco). Suspected colonies from HEA and XLD agar for Salmonella spp. and Shigella spp. were further confirmed by biochemical and serotyping using commercially available antisera (Denka-Seiken, Tokyo, Japan). Stool specimens were also inoculated onto MacConkey agar (Difco) plates and incubated overnight for identification of E. coli. Lactose fermenting colonies were further tested in pathogroup-specific PCR assays for identification of diarrhoeagenic E. coli. Stool specimens were also inoculated onto blood agar plates and kept under microaerophilic conditions for the presence of typical translucent colonies for presumptive identification of Campylobacter spp., which was subsequently confirmed by PCR assay.
Antimicrobial susceptibility testing
Antimicrobial susceptibility testing was performed using the disk diffusion method with commercially available disks. The disks used included ampicillin (AMP10), ceftriaxone (CRO30), chloramphenicol (C30), erythromycin (E15), gentamicin (GM10), nalidixic acid (NA30), ciprofloxacin (CIP5), ofloxacin (OFX5), norfloxacin (NOR10), meropenem (MEM30), streptomycin (STR10), azithromycin (AZM15), tetracycline (TET30), trimethoprim/sulfamethoxazole (SXT1.25/23.75), neomycin (N30) (Becton Dickinson, Sparks Glencoe, MD, USA) in accordance with the criteria recommended by Clinical and Laboratory Standards Institute [15]. According to CLSI guidelines, breakpoints for Enterobacteriaceae were used to determine antimicrobial susceptibility. Escherichia coli ATCC 25922 was used as a quality control strain.
PCR analysis
All the clinical isolates of V. cholerae were grown on Luria–Bertani (LB) Agar (Difco) plates. DNA extractions were carried out by phenol–chloroform extraction method [Reference Sambrook16] and were diluted to desired concentration for using as a template during PCR assay. Allele-specific PCR-based assay for the virulence-associated genes namely, cholera toxin subunit B (ctxB), toxin co-regulated pilus A (tcpA) and repeats-in-toxin A (rtxA) was performed using primer sets constructed by exploiting the single nucleotide polymorphism. DMAMA-PCR was done for identifying ctxB genotype 7 found in Haitian variant strains using primers ctxB-F3/Rv-Cla and for identifying genotype 1 found in classical biotype strains using ctxB-F4/Rv-Cla [Reference Naha17]. Another allele-specific PCR was carried out with the primer sets tcpA F1/El-Rev for El Tor type and tcpA F'2/El-Rev for variant type tcpA [Reference Raychoudhuri18]. Further, different alleles of rtxA were identified with one common forward (rtxAF) and two reverse primers (rtxA-R1 and rtxA-R2) specific for El Tor and rtxA-null mutant, respectively [Reference Ghosh19, Reference Ghosh20].
Pulsed-field gel electrophoresis
Pulsed-field gel electrophoresis (PFGE) was performed with the representative V. cholerae strains isolated during this period according to the Pulse Net protocol for V. cholerae [Reference Kam21]. DNA from Salmonella enterica serotype Braenderup strain H9812 was subjected to restriction digestion with XbaI (Takara, Japan) and employed as the universal size standard. The DNA of test strains was digested with NotI and subjected to electrophoresis. The gel was then stained for 30 min using ethidium bromide solution (5 µg/ml), followed by rinsing several times in distilled water. The band pattern was observed under UV illumination (BioRad, Hercules, CA, USA). The DNA fingerprint patterns of the test strains and the reference strains of V. cholerae were analysed using the computer software package BioNumerics (Applied Maths, Kortrijk, Belgium). The fingerprint patterns were also subjected to typing based on banding similarity and dissimilarity using the Dice similarity coefficient. The cluster analysis was performed based on the single-linkage method. Finally, the results were graphically represented as dendrograms.
Result
Confirmation of existence of an ongoing diarrhoeal outbreak
A total of 3003 diarrhoea cases were admitted to ID&BG Hospital during 1–15 August 2015 (Fig. 1), which was more than three times of the mean number of diarrhoea cases (911; s.d. = 111) admitted to the same hospital during the same period in the previous 7 years (2007–2014) confirming the occurrence of a diarrhoea outbreak in Kolkata municipal area during that period.
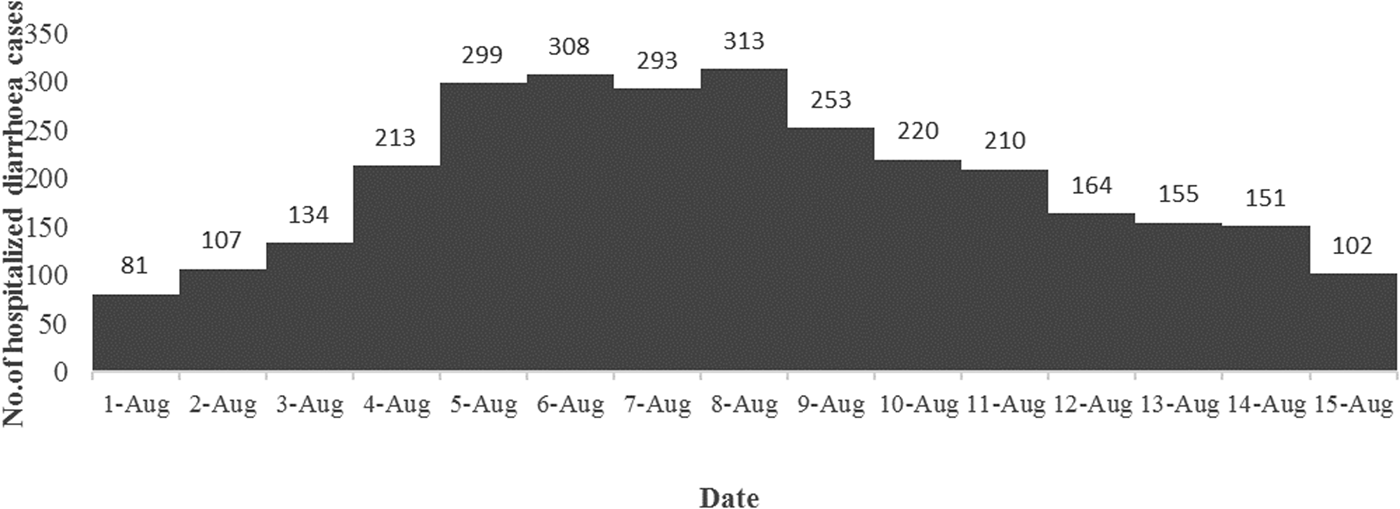
Fig. 1. Distribution of the number of diarrhoea cases admitted to the ID&BG Hospital, Kolkata during 1–15 August 2015.
Descriptive epidemiology
Among the total 3003 diarrhoea admitted cases during this period, 164 patients of all age groups and genders were recruited under the surveillance system following the sampling procedure stated earlier. All the age groups were affected in this outbreak (minimum age: 5 months; maximum: 99 years) and around a quarter of the cases (42 of 164) were below 5 years of age. Male patients predominated over females. Less than a fifth (~20%) of the patients had more than secondary education, which differed significantly between the two genders (male: 26.6%; female: 9.1%; P = 0.008). The average monthly income of the families was around Rs. 9000 (~US$135). Most of the families (>95%) used tap water for drinking purpose (Table 1).
Table 1. Socio-demographic characteristics of the diarrhoea cases

Spatial distribution of the diarrhoea cases
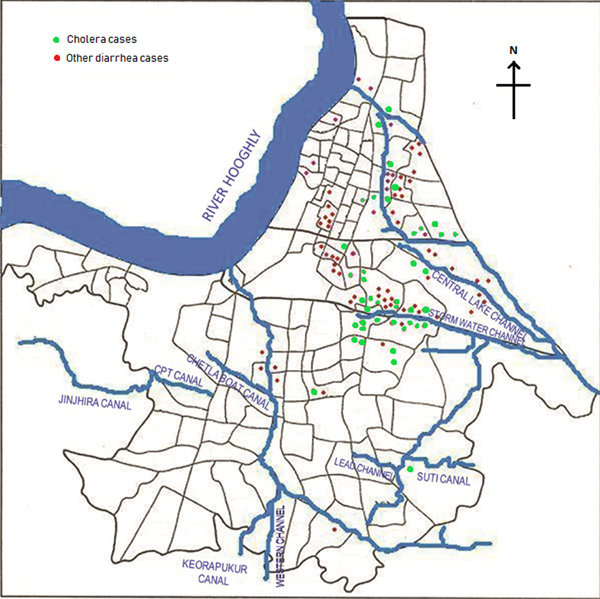
Of the total 164 diarrhoea cases recruited under the surveillance, 129 (79%) were residents of the Kolkata Municipal Corporation (KMC) area and the remaining patients came from neighbouring districts of Kolkata (mostly from north and south 24-Parganas). The KMC area is spread over 205 km2, and with a population close to 5 million residing in 144 wards (smallest administrative units of KMC), is dense (24 760 persons/km2) (https://www.kmcgov.in/KMCPortal/jsp/KMCPortalHome1.jsp). The river Hooghly runs along its western boundary and several canals traverse the municipal area for draining flood and storm waters into the river. Heavy rains and/or blockages of drainage systems for various reasons often lead water-logging in the affected areas in monsoon season, leading to contamination of drinking water and increasing the risk of waterborne diseases, especially gastro-intestinal infections. We plotted the diarrhoea cases occurring within the KMC area onto a map of that area to understand their spatial distribution and observed that most of the recruited 164 diarrhoea cases occurred either in areas along the canal which runs through Kolkata metropolis or within some selective geographic locations (Fig. 2). Distribution of cholera cases also revealed the similar patterns.
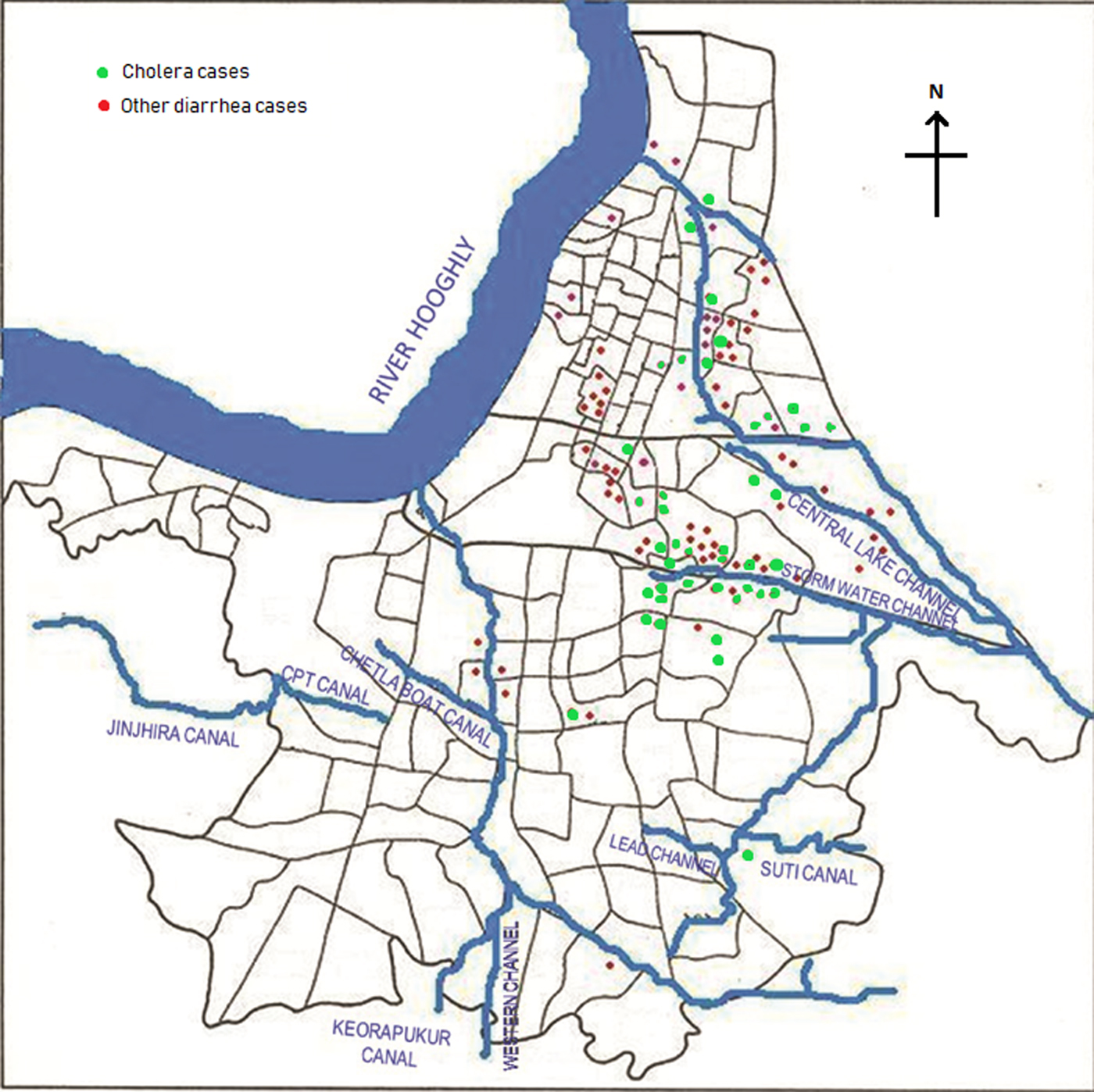
Fig. 2. Spot map of Kolkata Municipal Corporation area showing spatial distribution of diarrhoea cases during the outbreak. Green dots indicate culture-confirmed cholera cases whereas the red dots denote diarrhoea with other pathogens.
Clinical characteristics of the diarrhoea cases
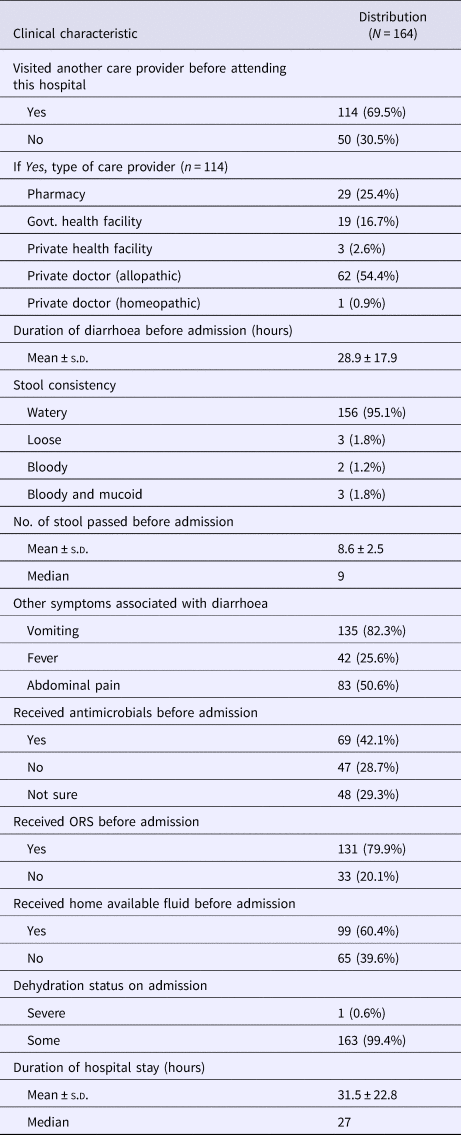
On an average, the patients attended the ID&BG Hospital more than a day after the onset of diarrhoea. About 70% of them had already visited a health care provider before attending the hospital – the commonest provider being an allopathic medical practitioner (54%), followed by a pharmacy outlet (25%) and a government health care facility (17%). Ninety-five per cent of the patients presented with watery diarrhoea, with a mean stool frequency of nine per day. All the recruited patients received intravenous fluid on admission and interestingly a majority of them received ORS or home available fluid for diarrhoea management even prior hospital admission. Forty-two per cent cases had received some antimicrobial drug(s) before admission, 30% could not tell about their intake of any antimicrobial agent. Except one patient with ‘severe’ dehydration, all others presented with ‘some’ degree of dehydration on admission (Table 2). One of these patients (a 52-year-old female with diarrhoea and vomiting) died in the hospital due to cardiovascular complication; remaining patients were improved during the hospital stay and discharged after complete recovery.
Table 2. Clinical characteristics of the diarrhoea cases
Laboratory investigation
Stool specimens were collected from all recruited diarrhoea patients and organisms were isolated from 80 (49%, 80/164) stool specimens. Out of the 80 microbiologically confirmed specimens, the most commonly isolated organism was V. cholerae O1 (50). From stool specimen of 28 patients, V. cholerae was isolated as the sole pathogen, whereas in 22 stool specimens, the organism was isolated along with other pathogen(s). Serological confirmation revealed that 49 belonged to O1, Ogawa and only one belonged to Inaba serotype. Patients aged between 18 and <30 years were mostly affected with cholera (13 of 27; 48%). Interestingly among 14 diarrhoea infants, five had cholera with V. cholerae O1 infection. No significant difference was found between male and female cases (M = 35%, F = 25%; P = 0.177) in isolation rate of V. cholerae organism.
Apart from diarrhoea cases recruited under the surveillance, forty additional stool specimens were collected from diarrhoea cases (not included in the surveillance) attending the hospital during the same period for better understanding of the aetiology of this outbreak and processed using similar procedures. However, the socio-demographic and clinical information was not collected for these cases.
The results obtained from testing a total of 204 stool specimen were as follows: microbiological analysis revealed that 63 (31%) were positive for V. cholerae O1, 23 (11%) for diarrhoeagenic E. coli, 14 (7%) for Shigella spp. and 13 (6%) with Campylobacter spp. In addition, six (3%) samples each were positive for V. fluvialis, five (2%) for V. cholerae non-O1, non-O139, two (1%) for Salmonella spp. and one (0.4%) was positive for V. parahaemolyticus. The distribution of total pathogens isolated from the outbreak samples is shown in Figure 3. A total of 98 stool specimens were positive for at least one pathogen and 73 samples were positive for sole pathogen. V. cholerae O1 was isolated as a sole pathogen from 40 (20%) of 204 stool samples screened.
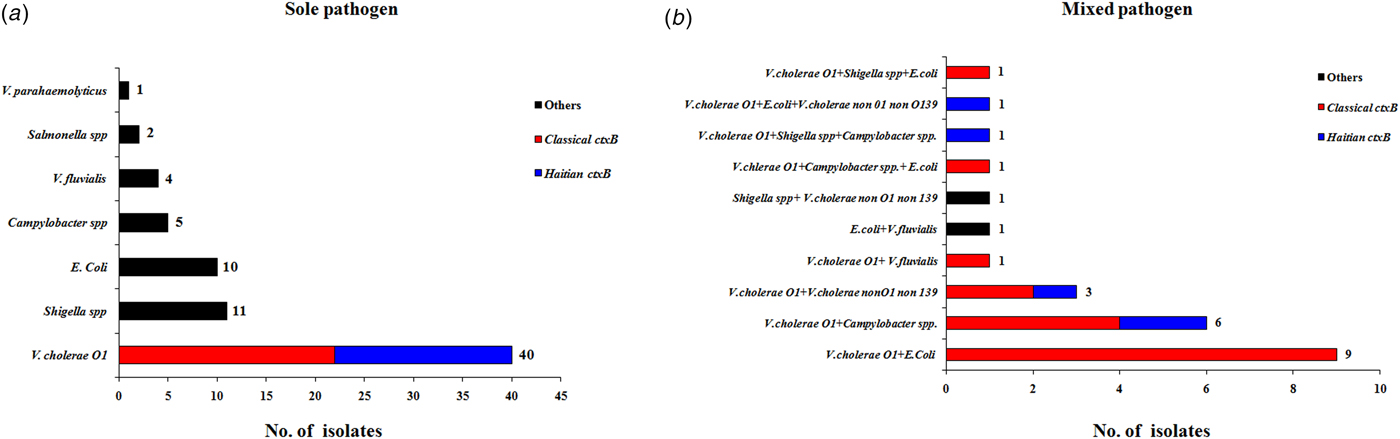
Fig. 3. Distribution of different enteric bacterial pathogens isolated from the stool specimens (N = 204) during 1–15 August 2015. V. cholerae strains were marked with two different colour codes based on their ctxB genetic status. (a) Indicates the distribution of sole pathogens in diarrhoeal patients and (b) depicts the number of patients infected with different set of mixed pathogens.
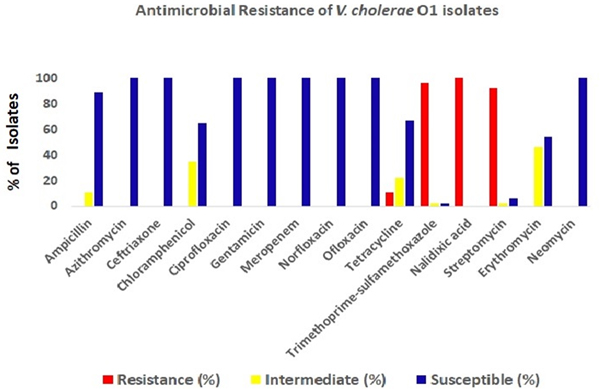
The antimicrobial disc diffusion test revealed that all the V. cholerae O1 isolates were resistant to nalidixic acid (100%), followed by co-trimoxazole (96%), streptomycin (92%) and tetracyclin (11%). But all isolates were sensitive to ciprofloxacin, ofloxacin, norfloxacin, meropenem, neomycin, gentamicin and azithromycin (Fig. 4).
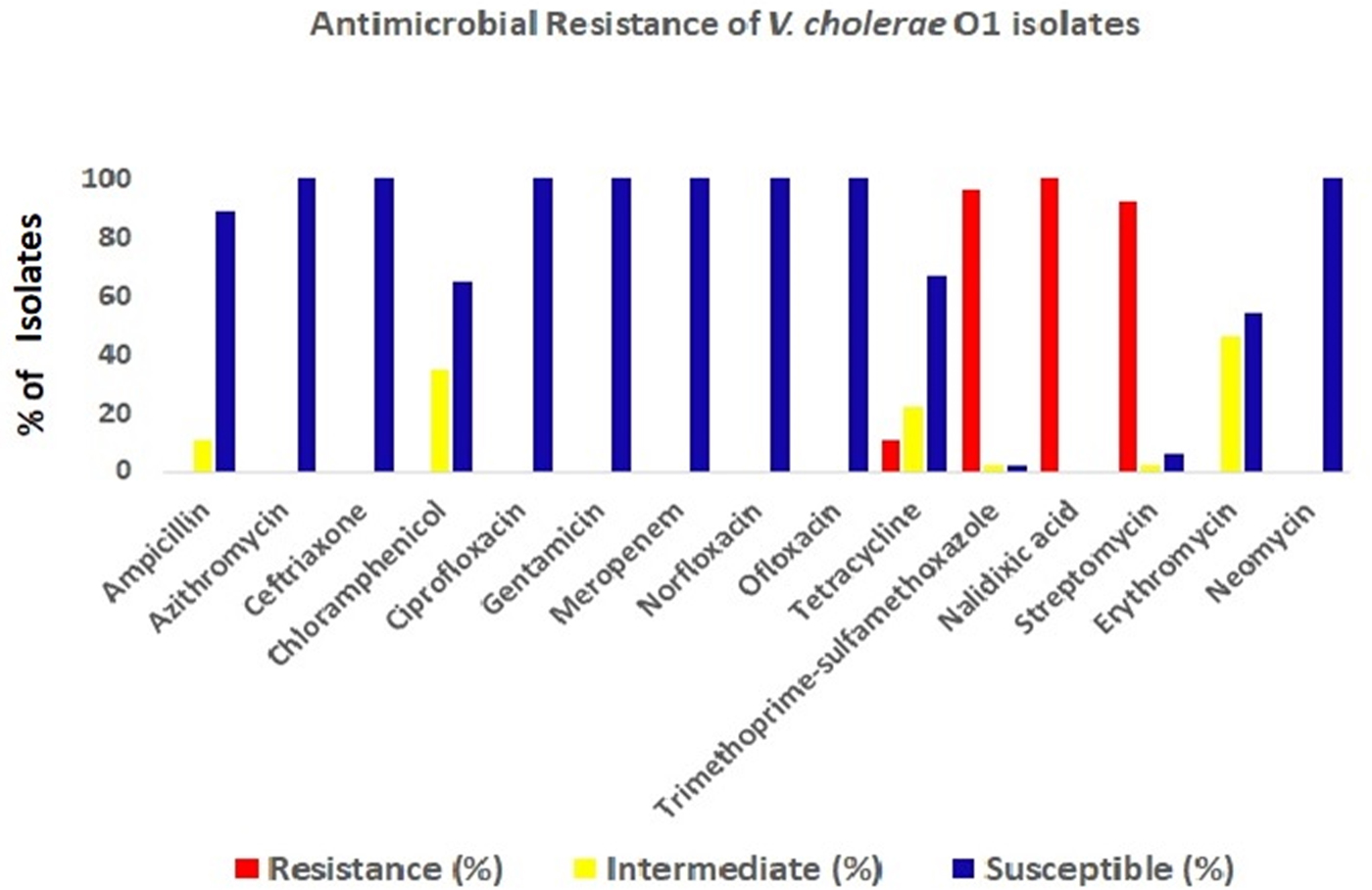
Fig. 4. Antibiotic-resistance profile of the V. cholerae isolates (n = 34) during the study period. Different colour codes for sensitive, resistance and intermediate level of resistance were used. Red, blue and yellow colour codes indicate resistant, sensitive and intermediate phenotypes, respectively.
Genetic features of the V. cholerae isolates
All the V. cholerae O1 isolates were tested by already established DMAMA PCR assay using two allele-specific forward primers and one common ctxB reverse primer for discriminating the classical, El Tor and Haitian type ctxB alleles. Surprisingly, around 65% of the V. cholerae strains yielded amplicons with ctxB classical-specific PCR primers and the rest of the isolates were positive for ctxB Haitian-specific PCR primer set indicating reappearance of classical ctxB (ctxB1) after 3 years of absence. Prior to this cholera upsurge, all the V. cholerae strains isolated at Kolkata contained the Haitian variant ctxB (ctxB7) from the middle of 2012 to first half of 2015. All the tested strains produced amplicons for Haitian-specific tcpA (tcpA CIRS) but not with the El Tor-specific tcpA. Interestingly, all the strains, which yielded amplicons for classical ctxB (ctxB1), were positive for El Tor-specific rtxA. On the other hand, the remaining strains, which produced amplicons for Haitian ctxB (ctxB7), were positive for Haitian-specific rtxA. So, based on the analysis of ctxB, tcpA and rtxA, the isolated strains during these 2 weeks period can be divided in two different genetic combinations namely, strains with Classical ctxB (ctxB1)_Haitian tcpA (tcpA CIRS)_El Tor rtxA and Haitian ctxB (ctxB7)_tcpA CIRS_variant rtxA (Table 3). To determine the genetic relatedness among V. cholerae strains, 14 representative strains were compared by using PFGE analysis. Dendrogram analysis using Bionumeric software (Applied Maths, Belgium) showed overall similarity of more than 95% and five pulsotype patterns of PFGE were obtained where predominant pulsotypes are P1 and P4. Interestingly, all the P1 pulsotype strains carried Haitian ctxB (ctxB7)_tcpA CIRS_variant rtxA, whereas strains containing P2-P5 pulsotypes had Classical ctxB (ctxB1)_Haitian tcpA (tcpA CIRS)_El Tor rtxA. But the isolation of V. cholerae strains with two genetic types from the affected population did not show any geographic clustering.
Table 3. Genetic background and mode of infection of V. cholerae O1 strains isolated in this study

*Mixed infection along with V. cholerae O1 includes other pathogens which have been depicted in Figure 3.
Discussion
In this study, V. cholerae was the most commonly isolated pathogen among the collected stool specimens. Little less than half of the recruited diarrhoea patients received antimicrobials before hospital admission, and interestingly, almost one-third of the recruited cases could not give any information on antimicrobial consumption history which possibly explains the absence of any pathogen in 84 collected stool specimens. Based on all these facts, we confirmed that there was an increased number of cholera cases during first 2 weeks of August 2015 in Kolkata metropolis area alongside the storm water channel and central lake channel which runs through east Kolkata. This might have occurred by contamination of drinking-water sources due to overflowing of canals/drains during the heavy rainfall in July 2015.
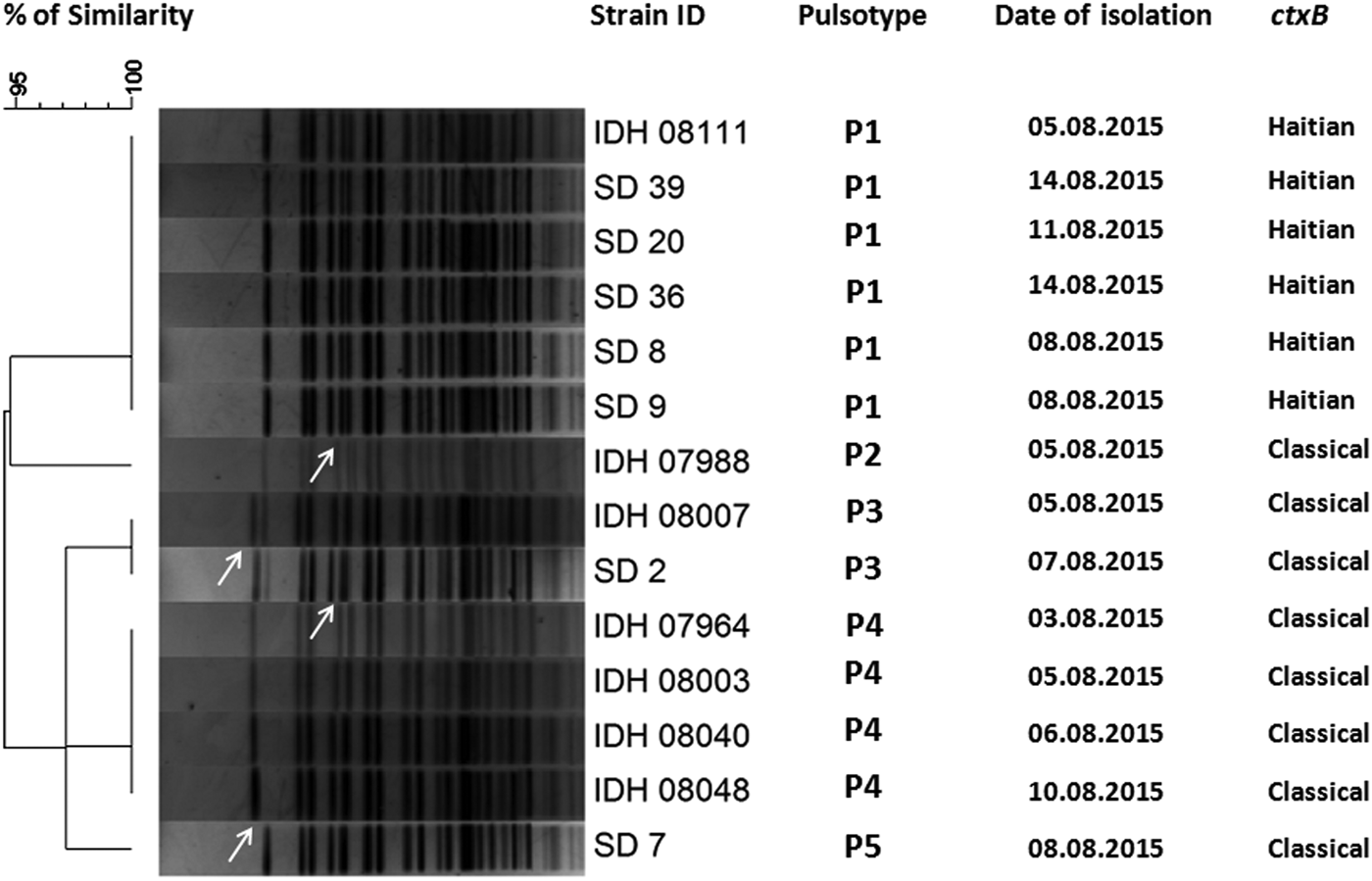
Fig. 5. Pulsed-field gel electrophoresis analysis of NotI-digested genomic DNA of V. cholerae strains (n = 14) isolated from diarrhoeal faecal samples in Kolkata, India during August 2015. The image of the gel was analysed using BioNumerics version 4.0 software (Applied Maths) based on the single-linkage method to generate the dendrogram. Per cent similarity was shown at the left-hand side. PFGE analysis revealed that all the isolates shared around 95% similarity along with five pulsotype patterns but P1 pulsotype strains carried Haitian ctxB whereas P2-P5 pulsotype strains contained classical ctxB.
Usually cholera epidemics are linked to monsoon followed by contamination of drinking water by sewage or polluted water bodies. Additionally, lack of proper distribution of safe drinking water, healthy hygiene and sanitation increases the burden in endemic areas [22]. Though in this study, majority of the recruited diarrhoea cases used municipal tap water supply as a drinking-water source, but instances of leakage in municipal pipeline water supply in West Bengal are common occurrence. Hence, chance of occurrence of waterborne disease outbreaks like cholera increases during monsoon or flood situations as drinking-water sources get easily contaminated by overflowing of sewerage drains [Reference Mridha23–Reference Datta25]. To avert such occurrences, piped water supply had been reinforced as improved drinking-water source, by Millennium development goal. Moreover, sustainable development goal 6 also addresses the issues of drinking water, sanitation and hygiene (WaSH), along with quality and sustainability of waterbodies worldwide [26] as periodic vigilance of, drainage and water supply system are the key to ensure supply of safe drinking water during natural calamities.
V. cholerae O1 was isolated from 31% of the stool specimens collected and processed from hospitalised patients. Genetic analysis of the V. cholerae strains revealed the reappearance of classical ctxB in Kolkata during 2015. Interestingly, 65% strains had genetic combination of classical ctxB, Haitian tcpA and El Tor rtxA. PFGE analysis of the representative V. cholerae strains yielded more than 95% similarity indicating the close genetic relatedness among these strains.
Cholera outbreaks due to multiple antibiotic-resistant V. cholerae have emerged as a major public health problem in last two decades [Reference Garg27]. In our study, multiple antibiotic-resistant V.cholerae isolates were recovered which was a matter of concern for a physician to treat the disease. In the present government-run routine disease surveillance system, there is no scope for keeping an account of the antibiotic sensitivity pattern of diarrhoea-causing pathogens, but it is immensely important as antimicrobial resistance has become a global public health threat.
This investigation had few limitations. Since, diarrhoea patients were recruited through ongoing hospital-based diseasesurveillance of ICMR – NICED, the actual number of affected people could not be ascertained, but still we could identify the situation when the cholera cases increased in Kolkata Metropolitan area and informed the local municipal authority about that. Second, we did not carry out any analytical epidemiological study in the field due to logistic constraints, but still from the literature review, it was evident that, contamination of water bodies was common in monsoon and post-monsoon period in Kolkata. We conclude that this outbreak in and around Kolkata during August 2015 was due to infection predominantly with V. cholerae O1, Ogawa and partially due to the other pathogens like Shigella, E. coli and Campylobacter. Isolation of multiple pathogens in the outbreak suggests that either remaining flood waters or the water supply are likely to be the source of the infections. However, confirmation would require testing the water supply or the remaining flood waters, which were not done during the episode.
Recommendations
Based on the findings, the local municipal corporation was alerted about the situation. Municipal corporation carried out information, education and communication activities in the affected areas to inform the residents of preventive measures like chlorination of drinking water, awareness generation on hand washing with soap and water for everyone after defecation and before preparing/serving/eating food, (ii) to create a proper drainage system in order to avoid stagnation of waste water, (iii) to boil the drinking water, (iv) not to buy antibiotics over the counter without consulting a physician for any diarrhoea episode in future. These measures will help to improve hygiene and infection control practices, which are needed for the prevention of diarrhoea.
Author ORCIDs
Shanta Dutta, 0000-0002-6897-7390
Acknowledgements
G.C. acknowledges the Postdoctoral fellowship received from the Okayama University. P.S. and R.N.S. acknowledge the CSIR fellowship (No. 09/482(0060)/2014-EMR-I) and ICMR fellowship (No. 3/1/3/JRF-2015/HRD-LS/88/40189/82) received from the Council of Scientific and Industrial Research, India and Indian Council of Medical Research, India, respectively.
Financial support
This study was supported in part by the Indian Council of Medical Research (ICMR), Government of India, Japan Initiative for Global Research Network on Infectious Diseases (J-GRID) of the Japan Agency for Medical Research and Development (AMED) under Grant Number JP18fm0108002 and National Institute of Infectious Diseases (NIID), Japan.
Conflict of interest
None.