Introduction
Amino acids (AA) are the physical basis of life as the substrates of protein synthesis in all tissues of animals. AA are divided into the essential, which cannot be synthesised by animals per se or the synthesis does not adequately meet the animal’s demand, or the non-essential for animals. The essential AA must be obtained in food and are comprised of arginine (Arg), cysteine (Cys), histidine (His), isoleucine (Ile), leucine (Leu), lysine (Lys), methionine (Met), phenylalanine (Phe), proline (Pro), threonine (Thr), tryptophan (Trp), tyrosine (Tyr) and valine (Val). The remaining AA are non-essential. Some essential AA can be classified as functional AA, and they not only serve as substrates for protein synthesis, but also function as signal molecules and participate in the regulation of metabolism, such as protein synthesis, of the body.
The synthesis of protein in tissues consists of sequential steps and consumes energy. The process of mRNA translation requires a substantial amount of cellular materials(Reference Shimizu, Inoue and Tomari1), and a sufficient supply of AA is a prerequisite for maintaining the optimal rate of protein synthesis(Reference Kimball2). These rate-limiting steps of biosynthetic processes are usually considered as the efficiency targets for regulatory mechanisms to control the processes(Reference Ma and Blenis3). For protein synthesis, the limiting steps are the initiation and elongation of a peptide chain. Increasing evidence has shown that some AA acts also as signalling molecules to change the phosphorylation status, then the functions of several proteins involved in the regulation of mRNA translation. Understanding these relationships between the functional AA and their regulation on mRNA translation could provide a base to develop nutritional approaches to improve protein synthesis in animal production.
There are two regulatory steps that limit the initiation of mRNA translation: one is the assembly of a ternary complex including eukaryotic initiation factors (eIF) 2 and Met-tRNA (Fig. 1), and the other is the assembly of the eIF 4F (eIF4F) complex in the mammalian target of rapamycin (mTOR) pathway (Fig. 2)(Reference Kimball2, Reference Ma and Blenis3). The ternary complex is composed of eIF2, GTP and Met-tRNA. The eIF4F complex contains three initiation factors: eIF4A, eIF4E and eIF4G(Reference Hershey, Merrick, Sonenberg, Hershey and Mathews4). After the complement of the assembly, both complexes bind to 40S ribosomal subunits(Reference Hinnebusch, Sonenberg, Hershey and Mathews5), which then scan toward the start codon and polypeptide formation begins. As for the elongation of peptide, the process is regulated by the activity of eukaryotic elongation factors (eEF). The initiation and elongation of protein synthesis can be affected by AA and several growth factors. The objective of the present review was to review the molecular mechanisms relating to AA regulation of protein synthesis, with particular focus on the effects of AA on the key signal pathways for the process of the protein translation initiation and elongation.

Fig. 1. Regulation of the assembly of the ternary complex by amino acids. eIF2, eukaryotic initiation factor-2; Met, methionine;?, unknown interactions between these factors; GCN2, general control nonderepressible 2; AA, amino acids. For a colour figure, see the online version of the paper.
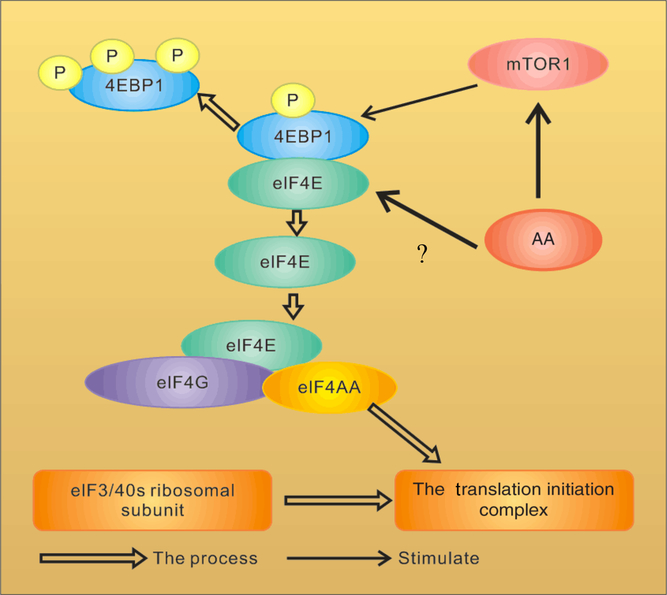
Fig. 2. Amino acid (AA) regulation of the assembly of eukaryotic initiation factor-4F (eIF4F) in the mammalian target of rapamycin (mTOR) pathway. 4EBP1, inhibitory 4E-binding protein-1;?, uncertain of whether AA regulate phosphorylation of 4EBP1 directly or indirectly through mTOR. For a colour figure, see the online version of the paper.
Amino acids regulate the assembly of the ternary complex
As illustrated in Fig. 1, the assembly of the ternary complex is a cyclic process starting from the binding of eIF2 to GTP and Met-tRNA, followed by the shift of the complex to 40S ribosomal subunits(Reference Kimball2, Reference Hinnebusch, Sonenberg, Hershey and Mathews5). When the start codon is recognised, active eIF2–GTP is converted to the inactive binary complex, eIF2–GDP, which is released to the cytoplasm. However, the assembly of the ternary complex requires more GTP–eIF2. The guanine nucleotide exchange reaction is mediated by eIF2B, which can be inactivated by the phosphorylation of eIF2α (subunit targeted for a few serine kinases that phosphorylate serine) through sequestering eIF2B from GDP as a competitive inhibitor(Reference Stipanuk6).
As eIF2 promotes the binding of the initiator Met-tRNA to 40S ribosomal subunits, the activity of which can be regulated by phosphorylation of itself, the phosphorylation status of eIF2α thus plays an important role in controlling the translation initiation, and this status can be regulated by some AA(Reference Hinnebusch, Sonenberg, Hershey and Mathews5, Reference Beugnet, Tee and Taylor7). Studies have shown that deprivation of AA enhanced eIF2α phosphorylation and inhibition of eIF2B activity, resulting in a decrease of protein synthesis in Ehrlich cells(Reference Scorsone, Panniers and Rowlands8, Reference Rowlands, Montine and Henshaw9). The eIF2α kinase, general control nonderepressible 2 (GCN2), has been identified to induce phosphorylation of eIF2α in mouse embryonic stem cells(Reference Berlanga, Santoyo and Haro10, Reference Sood, Porter and Olsen11) and can be activated by deprivation of AA(Reference Harding, Novoa and Zhang12). In addition, a minor increase in eIF2α phosphorylation was observed in CHO K1 cells when AA was withdrawn, suggesting that mammalian GCN2 may be activated over a period of AA withdrawal(Reference Beugnet, Tee and Taylor7), presumably through the accumulation of uncharged tRNA(Reference Hinnebusch13).
Despite a clear role of eIF2α phosphorylation in the regulation of protein synthesis by AA in vitro, results from animal studies in vivo have been less definitive(Reference Kimball2). When rats were fasted for 18 h and then fed a protein-containing meal, neither eIF2α phosphorylation nor eIF2B activity was affected by the meal in both the liver and skeletal muscle, although protein synthesis was increased by supply of the meal(Reference Yoshizawa, Kimball and Jefferson14). Oral administration of Leu increased protein synthesis in the skeletal muscle in food-deprived rats without causing any change in eIF2α phosphorylation or eIF2B activity(Reference Anthony, Anthony and Kimball15). However, in the livers of food-deprived young rats, lack of Trp, Leu or a mix of Leu, Ile and Val resulted in both an increase in eIF2α phosphorylation and an inhibition of eIF2B activity(Reference Anthony, Reiter and Anthony16). Recent results indicate that AA deprivation can regulate eIF2B activity independently of changes in eIF2α phosphorylation, probably via changes in phosphorylation of eIF2B itself(Reference Proud17). Another study showed that the addition of Ile, Leu, Met, and Thr together had no effect on eIF2α phosphorylation in mammary tissue of dairy cows(Reference Arriola Apelo, Singer and Lin18). Besides, removal of individual AA from the medium had no effect on eIF2α phosphorylation in mammary tissue slices and MAC-T cells, whereas depletion of all the essential AA increased eIF2α phosphorylation(Reference Appuhamy, Bell and Nayananjalie19, Reference Appuhamy, Knoebel and Nayananjalie20). Also, the addition of histidine, but not Leu or Met plus Lys reduced eIF2α phosphorylation in mammary glands of feed-deprived lactating cows(Reference Toerien, Trout and Cant21). Thus, AA affect protein synthesis through the regulation of the phosphorylation status of eIF2, as well as eIF2B.
Mechanism of regulating of binding proteins, mammalian target of rapamycin action and translation pathway
An important mechanism by which mTOR activity can be moderated is through interactions with other proteins. mTOR is a highly conserved protein kinase that regulates the growth and metabolism of mammals. The way mTOR regulates protein synthesis has been well described(Reference Ma and Blenis3, Reference Pazit and Michael22), as shown in Fig. 2. mTOR plays an important role in the second regulated step in the translation initiation involving the binding of mRNA to 40S ribosomal subunits, a reaction mediated by a triad of initiation factors collectively referred to as eIF4F(Reference Raught, Gingras, Sonenberg, Sonenberg, Hershey and Mathews23). The eIF4F complex is composed of three proteins: eIF4A, eIF4E (an RNA helicase that binds to the mGTP cap present at the 5′ end of mRNA)(Reference Beugnet, Tee and Taylor7) and eIF4G (a scaffolding protein that binds not only to eIF4A and eIF4E, but also to eIF3, which is bound to 40S ribosomal subunits)(Reference Kimball2). Thus, mRNA binds to 40S ribosomal subunits through the association of the eIF4F–mRNA complex with the eIF3-40S ribosomal subunit complex. The inhibitory 4E-binding protein-1 (4EBP1; also known as eIF4EBP1) inhibits eIF4G binding to eIF4E(Reference Ma and Blenis3) and the signal transduction-mediated phosphorylation of 4EBP1 leads to its dissociation from eIF4E, allowing the recruitment of eIF4G and eIF4A(Reference Gingras, Raught and Sonenberg24).
Studies have shown that AA affect the phosphorylation of 4EBP1. When HEK293 cells and CHO-IR cells were cultured in the media added with a mixture of AA, or individual AA, each of the essential AA was partially effective in promoting 4EBP1 phosphorylation and among them, Leu was the most potent one(Reference Hara, Yonezawa and Weng25, Reference Wang, Campbell and Miller26). In vivo experiments show similar results. In neonatal piglets(Reference Davis, Nguyen and Suryawan27) and rats(Reference Yoshizawa, Kimball and Jefferson14, Reference Anthony, Anthony and Kimball15, Reference Yoshizawa, Kido and Nagasawa28) subjected to overnight food deprivation, rates of protein synthesis were reduced in skeletal muscles and the liver, and feeding either a complete meal or a meal consisting of protein only rapidly reversed the inhibition, associated with enhanced phosphorylation of 4EBP1, dissociation of eIF4E from the inactive 4EBP1–eIF4E complex, and assembly of the active eIF4G–eIF4E complex.
Mode of action of mammalian target of rapamycin complex 1 and mammalian target of rapamycin complex 2
mTOR exists in two distinct multiprotein complexes, mTOR complex 1 (mTORC1) and mTOR complex 2 (mTORC2), which are known to differ in their subunit compositions, sensitivity to rapamycin and cell signalling activity(Reference Stefano, Kenneth and Stefano29, Reference Laplante and Sabatini30). Both complexes contain mTOR and mammalian lethal SEC13 protein 8; however, they are different in that mTORC1 contains the regulatory-associated protein of mTOR (raptor), whereas mTORC2 contains rapamycin-associated protein of mTOR (rapamycin-insensitive companion of TOR, or rictor)(Reference Winter, Fox and Kester31). With these differences present, mTORC1 is highly sensitive to rapamycin but mTORC2 is not. The mTORC1 activity is regulated by modulation of tumour suppressor tuberous sclerosis complex 1/2 (TSC 1/2) activity(Reference Laplante and Sabatini30, Reference Winter, Fox and Kester31). This complex is an active dimer that is purposed to limit mTORC1 signalling through the activity of GTPase. These proteins function to split a phosphate from GTP to yield GDP. Therefore, TSC 1/2 is known to negatively regulate mTORC1 activity by converting the GTPase protein Rheb into its inactive GDP-bound state(Reference Laplante and Sabatini30, Reference Jacobs, Goodman and Hornberger32). Thus, the GTPase activity of Rheb is highly governed by TSC 1/2. Moreover, when Rheb is in its active GTP-bound state, it translocates to the lysosome where mTORC1 is activated(Reference Winter, Fox and Kester31).
The mTORC1 pathway regulates cell growth in response to numerous cues, including AA, which promotes mTORC1 translocation to the lysosomal surface, its site of activation. The heterodimeric Ras-related GTP-binding protein (Rag)A/B–RagC/D GTPases, the regulator complex that tethers Rags to the lysosome, and the vacuolar-type H+-adenosine triphosphatase form a signalling system that is necessary for AA sensing by mTORC1. However, hepatitis B virus X-interacting protein and C7 or f59 as two additional regulator components form a pentameric complex that is essential for localising Rag GTPases to the lysosomal surface and activating mTORC1 in response to AA. In addition, to be a scaffold, the regulator promotes nucleotide exchange of RagB and of the highly related Rag A(Reference Bar-Peled, Schweitzer and Zoncu33). Rapamycin-insensitive mTORC1 phosphorylates and activates cell growth regulators S6 kinase 1 (S6K1) and S6 kinase 2 (S6K2) and activates ribosomal protein S6(Reference Holz, Ballif and Gygi34), and it phosphorylates and inactivates the translational repressors eukaryotic translation initiation factor 4EBP1; further, 4EBP1 targets and prevents eIF4E–eIF4G interaction and thus inhibits translation initiation(Reference Richter and Sonenberg35). mTOR inhibitor rapamycin dephosphorylates S6K1 and 4EBP1, leading to G1 growth arrest and inhibition of ribosome biosynthesis and autophagy(Reference Easton and Houghton36, Reference Inoki, Ouyang, Li and Guan37). The anti-tumour activity of rapamycin as a single agent has been described in vitro and in vivo (Reference Dilling, Dias and Shapiro38, Reference Noh, Mondesire and Peng39). Furthermore, mTORC2 activates protein kinase B (PKB, also known as Akt) and regulates the actin cytoskeleton(Reference Jacinto, Loewith and Schmidt40, Reference Sarbassov, Ali and Kim41). Rapamycin dephosphorylates the mTORC2 component rictor(Reference Akcakanat, Singh and Hung42). Although rictor phosphorylation may affect Akt phosphorylation, the biological consequence of its phosphorylation is relatively unknown(Reference Dibble, Asara and Manning43). Both mTORC1 and mTORC2 respond to hormones and growth factors(Reference Wullschleger, Loewith and Hall44); however, only mTORC1 has been proposed to be acutely regulated by nutrients such as AA(Reference Dann, Selvaraj and Thomas45, Reference Huang, Dibble and Matsuzaki46). Many reports have suggested that AA activate mTORC1, increase cell growth through increased ribosome biogenesis and protein biosynthesis and suppression of autophagy(Reference Wullschleger, Loewith and Hall44, Reference Kimball and Jefferson47). Among AA, Leu is in the primacy as discussed below. Growth factor signals have little or no effect on mTORC1 signalling in the absence of AA(Reference Hara, Yonezawa and Weng25).
Target of rapamycin interrelating proteins and regulation pathway of mammalian target of rapamycin signalling
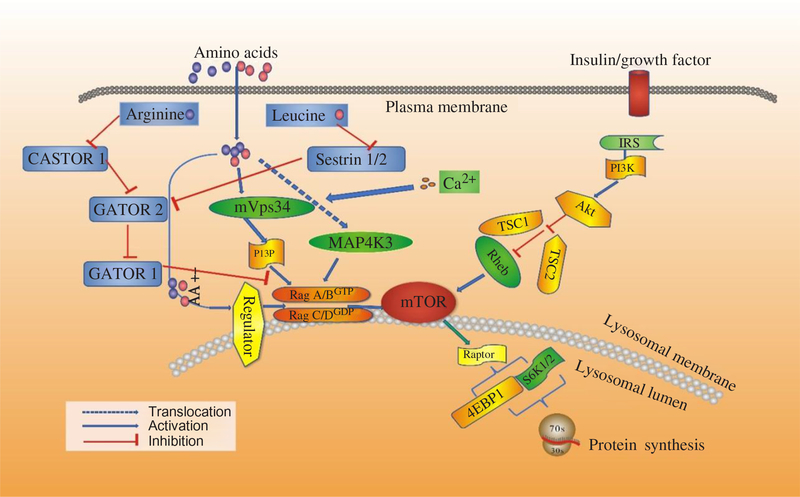
Recent studies have shown that AA regulate phosphorylation of 4EBP1 indirectly, and three new proteins have been revealed to mediate the regulatory effects on protein synthesis by AA, including the Ragulator–Rag complex, vacuolar protein sorting 34 (Vps34) and mitogen-activated protein kinase 3 (MAP4K3). Then mTORC1 regulates cell growth by coordinating upstream signals from growth factors, intracellular energy levels, and AA availability(Reference Guertin and Sabatini48). The tuberous suppressor complex, TSC1/TSC2, affects the activity of mTORC1 by regulating the GTP-loading state of Rheb (Ras homologue enriched in brain), a Rheb-related GTP binding protein, and GTP–Rheb interacts with and activates mTORC1(Reference Laplante and Sabatini49). However, TSC1/TSC2 is dispensable for the regulation of mTORC1 by AA, and in cells lacking TSC2 the mTORC1 pathway is sensitive to AA starvation but resistant to growth factor withdrawal(Reference Smith, Finn and Tee50, Reference Roccio, Bos and Zwartkruis51). Recent studies have shown that Rag GTPases, which are also members of the Ras family of GTP-binding proteins, are AA-specific regulators in the mTORC1 pathway(Reference Kim, Goraksha-Hicks and Li52, Reference Sancak, Peterson and Shaul53). Mammals express four Rag proteins (RagA, B, C and D), which form heterodimers consisting of RagA or RagB with RagC or RagD. RagA and RagB, as well as RagC and RagD, are highly like each other and are functionally redundant(Reference Sancak, Peterson and Shaul53, Reference Sekiguchi, Hirose and Nakashima54). Further, GTP–RagB interacts with mTORC1 by binding to raptor (regulatory-associated protein of mTOR), one part of the mTORC1 complex(Reference Shaw55), and AA induce the mTORC1–Rag interaction by promoting the loading of RagB with GTP(Reference Sancak, Peterson and Shaul53). The activation of the mTORC1 pathway by AA correlates with the movement of mTORC1 from an undefined location to a compartment containing Rab7(Reference Sancak, Peterson and Shaul53), a marker of both late endosomes and lysosomes(Reference Chavrier, Parton and Hauri56, Reference Luzio, Pryor and Bright57). Sancak et al.(Reference Sancak, Peterson and Shaul53) pointed out that AA stimulate the translocation of mTORC1 to the lysosomal surface, where the Rag GTPases reside irrespective of their GTP-loaded states or AA availability, and the translocation of mTORC1 to lysosomes does not depend on growth factors, Rheb or mTORC1 activity(Reference Yasemin, Liron and Roberto58). Moreover, a trimeric regulator complex, including MP1, p14, and p18, is mostly found to interact with Rag GTPases and colocalises with them on lysosomal membranes and is necessary for the AA-dependent recruitment and activation of mTORC1 (Fig. 3)(Reference Yasemin, Liron and Roberto58, Reference Deldicque, Theisen and Francaux59). In addition to the Ragulator–Rag complex, other components have also been implicated in the upstream control of mTORC1 by AA(Reference Proud17). One of them is a homologue of the mammalian protein kinase, MAP4K3, which is identified as a nutrient-sensitive regulator of mTOR signalling by a recent RNA interference screen in Drosophila cells(Reference Findlay, Yan and Procter60).
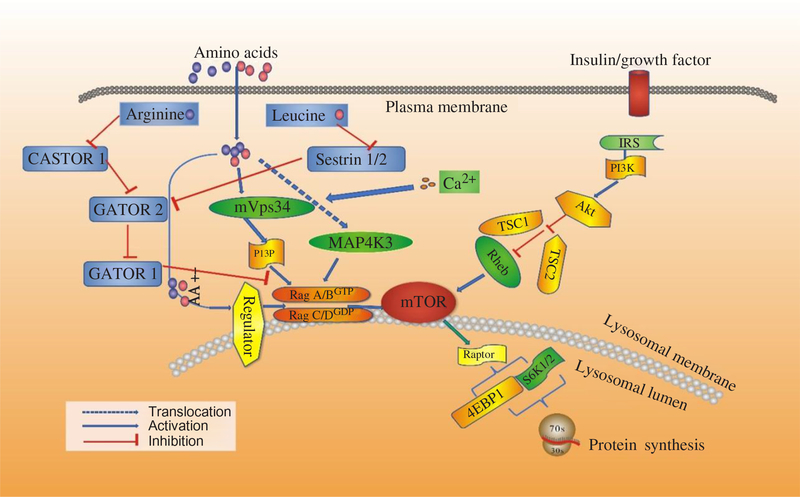
Fig. 3. Signalling by amino acids (AA) and other factors, target of rapamycin (TOR) interrelating protein activation and mammalian target of rapamycin (mTOR) signalling for protein synthesis. 4EBP1, inhibitory 4E-binding protein-1; Akt, protein kinase B; IRS, insulin receptors; MAP4K3, mitogen-activated protein kinase-3; PI3K, phosphatidylinositol-3-kinases; PI3P, phosphatidylinositol-3-phosphate; Rag, Ras-related GTP-binding protein; Rheb, Ras homologue enriched in brain; S6K1, ribosomal protein S6 kinase β-1; TSC1/TSC2, tuberous suppressor complex; Vps34, vacuolar protein sorting-34. For a colour figure, see the online version of the paper.
Another possible mediator of AA signalling to mTOR is the class III phosphotidyl-inositol 3-kinase mVps34(Reference Byfield, Murray and Backer61, Reference Gulati, Gaspers and Dann62). AA induce an extracellular Ca influx that activates calmodulin, which in turn binds to and activates mVps34, and mVps34 then produces phosphatidylinositol-3-phosphate (PI3P) that somehow activates mTORC1. The mechanism also involves the formation of a calmodulin–mVps34–mTORC1 super-complex(Reference Gulati, Gaspers and Dann62). Additionally, Vps34 has also been reported to regulate Rab7-mediated vesicular trafficking(Reference Stein, Yan and Karen63, Reference Kinchen, Kimon and Johann64). The product of hVps34 and PI3P is involved in recruiting proteins to endosomal membranes containing FYVE or phox homology (PX) domains(Reference Lemmon65), with these PI3P-rich microdomains serving as platforms to build signalling complexes(Reference Miaczynska, Pelkmans and Zerial66, Reference Maya, James and Jonathan67). Given the Rag-dependent re-localisation of mTORC1 to Rag7-positive structures, this connection could provide a potential explanation for how Vps34 depletion could interfere with the AA control of mTORC1 activity(Reference Findlay, Yan and Procter60). Recent studies have revealed the functions of class III PI3 kinase in murine myotubes(Reference MacKenzie, Hamilton and Murray68), HepG2, and CHO cells(Reference Stein, Yan and Karen63). In states of AA starvation, hVps34 demonstrated reduced function. However, the functions restored with re-addition of AA(Reference Gulati, Gaspers and Dann62, Reference Stein, Yan and Karen63). Further, for the first time, mTOR signalling was enhanced in response to an acute stimulation with the proteinogenic AA, Leu, in cultured human myotubes, and hVps34 served as a nutrient-sensing protein for mTOR signalling mediated by AA(Reference Petra and David69). Moreover, hVps34 plays an unappreciated role in nutrient sensing and activation of ribosomal protein S6 kinase β-1 (S6K1) and is a component of the AA-regulated arms of S6K1 regulation(Reference Maya, James and Jonathan67). Many advances have been made to illuminate the mechanism by which AA regulate mTORC1. The Rag complex, MAP4K3 and mVps34 have been implicated. Some studies showed that MAP4K3 converged on the signalling through mTOR in mammalian cells by activating proteins Rag 1-4 and class III phosphatidylinositol 3′-kinase Vps34(Reference Sancak, Peterson and Shaul53, Reference Byfield, Murray and Backer61, Reference Gulati, Gaspers and Dann62). A recent study suggests that it is the activity of Vps34 that is the primary modulator of the Leu-stimulated mTOR signalling in rat muscle(Reference Maya, James and Jonathan67), which is different from the interaction between MAP4K3 and Vps34 as described previously. More studies should be done to verify where these proteins fit into the AA regulatory pathway.
Amino acids regulate the elongation process of protein synthesis
Ribosomes join AA together in the sequence determined by the mRNA to make the corresponding protein, and AA are brought onto ribosomes attached to tRNA. Further elongation requires the help of elongation factor proteins and tRNA with attached AA (called aminoacyl tRNA) that are brought onto ribosomes by one such elongation factor(Reference Arriola Apelo, Singer and Lin18). This factor is termed EF-Tu in prokaryotes and EF1 in eukaryotes. To complete the round of elongation, a second elongation factor, called EF-G in prokaryotes and eukaryotic elongation factor 2 in eukaryotes, is needed. However, ribosomes have three aminoacyl tRNA binding sites: the acceptor site (A), the peptidyl site (P) and the exit site (E)(Reference Carlberg, Nilsson and Nygard70). This elongation factor moves tRNA such that the spent tRNA that has lost the protein chain moves fully into the E site, and tRNA with the growing protein chain moves fully into the P site. The mRNA is also shifted over one codon by EF-G so that the next codon is in the A site. The A site is now empty of tRNA and the next aminoacyl tRNA can be brought into it.
AA modulate the translation elongation machinery through the control of the activity of eEF2. The eEF2 is subject to phosphorylation, and the phosphorylated form of the protein is inactive in translation(Reference Carlberg, Nilsson and Nygard70). AA depletion can result in increased phosphorylation of eEF2(Reference Wang, Campbell and Miller26). In addition, Arriola Apelo et al.(Reference Arriola Apelo, Singer and Lin18) revealed the effects of Leu, Ile, Met and Thr and their interactions on the activity of eEF2 in mammary tissue. According to this study, eEF2 phosphorylation was linearly decreased by Ile and Leu, Thr caused a curvilinear decrease in eEF2 phosphorylation, and both Ile and Leu negatively interacted on eEF2, and Thr tended to inhibit the Leu effects on eEF2. Furthermore, mTORC1 could control the phosphorylation of eEF2(Reference Proud17), and the phosphorylation of eEF2 by eEF2K, a calmodulin-dependent kinase downstream of mTOR and S6K1, also inhibits its translation elongation activity(Reference Arriola Apelo, Singer and Lin18).
Arginine and leucine in the primacy on mammalian target of rapamycin complex 1 signalling and synthesis of protein
Although several AA can regulate mTORC1 signalling through the Rag GTPase, Arg and Leu are the best established and deprivation of either strongly inhibits mTORC1 in various cell types(Reference Hara, Yonezawa and Weng25). A complex which is called GATOR can interact with the Rags and is composed of two subcomplexes (GATOR1 and GATOR2). Inhibition of GATOR1 subunits makes mTORC1 signalling resistant to AA deprivation, while inhibition of GATOR2 subunits suppresses mTORC1 signalling(Reference Bar-Peled, Chantranupong and Cherniack71). The Leu-binding capacity of Sestrin2 is required for Leu to activate mTORC1 in cells. This indicates that Sestrin2 is a Leu sensor for the mTORC1 pathway(Reference Wolfson, Chantranupong and Saxton72). However, stimulation by Arg in cells to activate mTORC1 is not well understood. Moreover, CASTOR1 works as an Arg sensor for the mTORC1 pathway and interacts with GATOR2 and is required for Arg deprivation to inhibit mTORC1(Reference Chantranupong, Scaria and Saxton73). CASTOR1 homodimerises and can also heterodimerise with the related protein CASTOR2(Reference Chantranupong, Scaria and Saxton73). Arg disrupts the CASTOR1–GATOR2 complex by binding to CASTOR1. The proteins that sense AA and signal to the Rag GTPases were indefinable until recently. Sestrin2 as a cytosolic Leu sensor and solute carrier family 38 member 9 (SLC38A9) as a putative lysosomal Arg sensor for the mTORC1 pathway was identified(Reference Wolfson, Chantranupong and Saxton72, Reference Wang, Tsun and Wolfson74). While Sestrin2 interacts with GATOR2 to inhibit mTORC1 signalling in the absence of Leu, SLC38A9 forms a super complex with a regulator and is necessary for transmitting Arg, but not Leu, sufficiency to mTORC1(Reference Wolfson, Chantranupong and Saxton72, Reference Wang, Tsun and Wolfson74, Reference Chantranupong, Wolfson and Orozco75). For Leu to activate mTORC1, Sestrin2 must be able to bind Leu and work as a Leu sensor for the mTORC1 pathway. Furthermore, Sestrin2 has an appreciable affinity for Met. It would not be surprising if, in contexts where Leu concentrations are low and those of Met are high, Sestrin2 may serve as a Met sensor for the mTORC1 pathway (Fig. 3)(Reference Wolfson, Chantranupong and Saxton72, Reference Chantranupong, Wolfson and Orozco75).
The primacy of Leu on mTORC1 signalling and the effect on protein synthesis is of great interest, nutritionally both in terms of the reduction of loss of lean body mass in various disease states and in the maintenance/enhancement of lean body mass in the healthy condition. Specifically, Leu has been reported to activate the mTOR/S6K1 signalling pathway(Reference Kimball, Shantz and Horetsky76–Reference Lee, Jo and Lee78). Leu, acting through an mTORC1-dependent pathway, stimulates the translation of specific mRNA both by increasing availability of eIF4E and by stimulating phosphorylation of S6(Reference Karlsson, Nilsson and Nilsson79). In addition, Leu induces cell migration and Rac activation controlling cytoskeletal reorganisation through mTORC2(Reference Hernández-Negrete, Carretero-Ortega and Rosenfeldt80). However, deprivation of Leu leads to a cellular response overlapping with but distinct from mTOR inhibition(Reference Peng, Golub and Sabatini81), and deprivation and rapamycin have been proposed to have synergistic effects on protein synthesis(Reference Mordier, Deval and Bechet82).
The synthesis of protein in tissues is rapidly stimulated after oral intake of nutrients, and Leu appears to mediate most of the effective AA on protein metabolism. The central role of Leu mediating the anabolic effects of protein/AA on muscle protein metabolism was noted from the mid-1970s(Reference Buse and Reid83, Reference Tischler, Desautels and Goldberg84). Not only does Leu stimulate protein synthesis in skeletal muscle, but also affects protein degradation in skeletal muscle and the liver, through TORC1-dependent and independent mechanisms. Protein synthesis is associated with an increase in 4EBP1 phosphorylation, a decrease in association of 4EBP1 with eIF4E, an increase in association of eIF4G with eIF4E, and an increase in S6K1 phosphorylation. Long et al.(Reference Long, Ortiz-Vega and Lin85) showed that binding of Rheb to mTOR was promoted by the presence of AA and inhibited by the withdrawal of a mixture of AA or Leu only, but it seemed unlikely that this mechanism could account for the specific physiological effects of Leu in intact animals. Another unique signalling function of Leu in the regulation of metabolism, in part through the regulation of mTOR signalling, has been extensively supported by studies in vivo, primarily in rodents(Reference Stipanuk6) and in human subjects(Reference Coffey and Hawley86, Reference Vary and Lynch87).
Leu also acts on the central nervous system to control overall food intake through TORC1, and food selection probably through GCN2(Reference Hao, Sharp and Ross-Inta88, Reference Maurin, Jousse and Averous89). Direct administration of l-Leu (but not l-Val) near the arcuate nucleus region in rat hypothalamus stimulates hypothalamic TOR signalling and results in decreased food intake (anorexia) and significant weight loss(Reference Cota, Proulx and Smith90). Activation of GCN2 occurs in response to a deficiency of any essential AA; however, the regulation of mTORC1 is most responsive to specific individual AA(Reference Stipanuk6). Among AA, Leu and Arg share nearly comparable inhibitory effects(Reference Roh, Han and Tzatsos91) but underlying mechanisms are puzzling; other than their participation in polypeptide chain elongation through the mediation of six tRNA, these two AA have no commonalities in their transport or metabolism-regulatory functions. Additionally, it should be mentioned that alterations in the extracellular concentration of AA whose transport is Na+ linked, for example, glutamine but (not Leu), will lead to parallel alterations in cell hydration; cell swelling induced by any mechanism is accompanied by activation of several anabolic pathways, including mTORC1, whereas cell dehydration is inhibitory(Reference Schliess, Richter and Vom92, Reference Krause, Bertrand and Maisin93).
Leu also interacts with carbohydrate on metabolism in cells and the body. Leu plus carbohydrate have the same effect as Leu alone, and administration of either Ile or Val alone had no effect and administration of Leu produced only a slight, transient rise in plasma insulin concentration at 30 min, whereas carbohydrate caused a much greater increase that was maintained at 60 min(Reference Anthony, Lang and Crozier94). Using a meal-feeding model, Lynch et al.(Reference Lynch, Patson and Anthony95) confirmed the stimulatory effect of Leu on tissue protein synthesis in gastrocnemius, kidney and adipose tissue, whereas a carbohydrate meal had no effect despite a robust increase in plasma insulin. Further, Leu also promotes leptin synthesis in the adipocyte(Reference Lynch, Gern and Lloyd96), and both Leu and leptin action on leptin-sensitive neurons, signals that reflect immediate and long-term adequate nutrition, activate TORC1 to suppress further food intake. The stimulation of TORC1 by Leu appears to be initiated at an intracellular site. In Xenopus oocytes, extracellular Leu is unable to promote S6K1 phosphorylation, but expression of a recombinant system L transporter confers responsiveness to extracellular Leu and direct intracytoplasmic injection of l-Leu (or Trp, Arg, Lys and glycine, but not d-Leu, alanine, Pro, glutamate or glutamine) stimulates S6K1-P in a rapamycin-sensitive manner(Reference Christie, Hajduch and Hundal97).
Leu and Phe both promoted synthesis or regulated secretion of α-amylase secretion in dairy goats(Reference Yu, Xu and Liu98). Phe also regulates the synthesis of trypsin and lipase through mRNA translation initiation factors S6K1 and 4EBP1(Reference Guo, Tian and Shen99). The branched-chain AA Val, Leu, and Ile are essential AA; among these, Leu particularly stimulates protein synthesis(Reference Liu, Liu and Liu100). It has been found that Leu specifically stimulates phosphotidyl-inositol 3-kinase(Reference Tesseraud, Bigot and Taouis101), the kinase for mTOR(Reference Yoshizawa, Hirayama and Sekizawa102), resulting in stimulation of the protein synthesis machinery, including ribosomal S6 kinase and 4E binding protein in skeletal muscles(Reference Um, D’Alessio and Thomas103). However, suppression of proteolysis by Leu does not involve an mTOR signalling pathway in mouse C2C12 myotubes(Reference Mordier, Deval and Bechet82) and isolated rat hepatocytes(Reference Kanazawa, Taneike and Akaishi104). The suppression of myofibrillar proteolysis by Leu does not involve the mTOR signalling pathway in chick myotubes cultured in vitro, and oral administration of Leu suppresses myofibrillar proteolysis in chicks in vivo (Reference Nakashima, Ishida and Yamazaki105).
Conclusion
AA play a key role in both maintaining body metabolism and signalling protein synthesis. Several new proteins have been found in the AA-regulating pathways, and their regulatory mechanisms remain unclear. Future studies will need to depict these mechanisms because of their critical roles in modulating protein synthesis and other metabolism in the body.
Author ORCID
Yangchun Cao 0000-0003-1033-2909
Acknowledgements
The present review was supported by the National Natural Science Foundation of China (grant no. 31472122 and 31672451) and the Collaborative Innovation Major Project of Industry, University, Research, and Application in the Yangling Demonstration Zone (grant no. 2016CXY-18). The paper represents the original work of the authors. Y. C. was responsible for researching, writing and preparing the manuscript. K. L. and I. H. R. A. contributed the figures. S. L., C. C. and J. Y. provided intellectual input, proof reading and corrections. The authors declare that they have no conflicts of interest.