Introduction
Cowpea [Vigna unguiculata (L.) Walp.] is an important legume grown in the tropical and subtropical regions of Africa, Asia, South America, and southern parts of the United States and Europe (Singh et al. Reference Singh, Raj, Dashiell and Jackai1997). In addition to producing grain, cowpea is a key source of fodder, and its leaves and pods are consumed as green vegetables. Cowpea is highly tolerant to drought and heat and has consequently been recognized as an important component of food security in sub-Saharan Africa. However, yields are constrained by abiotic stresses, pests, and diseases. One of the most important constraints of cowpea is the plant parasite cowpea witchweed [Striga gesnerioides (Willd.) Vatke].
Striga gesnerioides is a major contributor to cowpea yield losses throughout sub-Saharan Africa, India, and Southeast Asia (Parker and Riches Reference Parker and Riches1993). The parasite is difficult to control, with each plant producing as many as 200,000 seeds, contributing to recalcitrant seedbanks capable of persisting for decades (Bebawi et al. Reference Bebawi, Eplee, Harris and Norris1984; Parker and Riches Reference Parker and Riches1993). A survey of 153 cowpea fields across six countries in West Africa found S. gesnerioides among 40% of the sampled locations, and as many as 81% of cowpea fields were infested in northeast Nigeria (Cardwell and Lane Reference Cardwell and Lane1995; Dugje et al. Reference Dugje, Kamara and Omoigui2006). Under severe infection, complete yield loss may occur (Emechebe et al. Reference Emechebe, Singh, Leleji, Atokple and Adu1991). The degree of Striga infestation varies depending on soil fertility and cropping system and is generally most severe in depleted, sandy soils in monocrop systems with few crop rotations (Cardwell and Lane Reference Cardwell and Lane1995).
Striga gesnerioides is part of the Orobanchaceae family. Mohamed et al. (Reference Mohamed, Musselman and Riches2001) characterized 28 Striga species and six subspecies, among which purple witchweed [Striga hermonthica (Delile) Benth.], Asiatic witchweed [Striga asiatica (L.) Kuntze], and S. gesnerioides are the most economically important. Striga hermonthica and S. asiatica primarily infect cereals in the Poaceae family, while the primary hosts of S. gesnerioides are cowpea and wild legume species. However, S. gesnerioides is capable of parasitizing hosts across genera including Ipomea, Jaquemontia, Merremia, Euphorbia, and Nicotiana (Mohamed et al. Reference Mohamed, Musselman and Riches2001). Several different Striga biotypes associated with resistance breakdown have been identified (Gethi et al. Reference Gethi, Smith, Mitchell and Kresovich2005). Breakdown of resistance has been reported in both Tanzania and West Africa (Doggett Reference Doggett1952; Ramaiah Reference Ramaiah and Musselman1987). A deeper understanding of the diversity among these important plant parasites, the effectiveness of various host resistance genes, and the progression of population structure is vital information for the development of elite cowpea varieties with durable resistance.
Several studies have examined genetic diversity among Striga species, and associations between genetic distance and host specificity were typically low. Genetic analysis of S. hermonthica and S. asiatica populations from Kenya, which were genotyped using amplified fragment-length polymorphism (AFLP) markers, indicated small genetic distances between populations, averaging 0.032 in S. asiatica and 0.015 in S. hermonthica (Gethi et al. Reference Gethi, Smith, Mitchell and Kresovich2005). Marginal levels of population structure were found in the autogamous species, S. asiatica, and no significant structure was observed among populations of the obligate outcrosser, S. hermonthica (Gethi et al. Reference Gethi, Smith, Mitchell and Kresovich2005). Similarly, differentiation was low between S. hermonthica populations collected from grain sorghum [Sorghum bicolor (L.) Moench ssp. bicolor] and millet (Pennisetum spp.) in Burkina Faso and Sudan (Bharathalakshmi et al. Reference Bharathalakshmi, Werth and Musselman1990). Interestingly, greater geographic differentiation was found between populations from Burkina Faso and Sudan than between host-specific populations collected within Burkina Faso (Bharathalakshmi et al. Reference Bharathalakshmi, Werth and Musselman1990). Low genetic divergence was also reported between S. hermonthica populations collected from sorghum, millet, maize (Zea mays L.), and wild grasses across Burkina Faso, Mali, and Niger (Olivier et al. Reference Olivier, Glaszmann, Lanaud and Leroux1998). While moderate differentiation was found among S. hermonthica populations collected from Ethiopia, genetic differences were not associated with variable host specificity (Welsh and Mohamed Reference Welsh and Mohamed2011). Similar differentiation was identified between S. hermonthica collected from Nigeria and Kenya. However, populations collected from rice (Oryza sativa L.) were genetically distinct from populations collected from maize and sorghum despite their close geographic proximity (Unachukwu et al. Reference Unachukwu, Menkir, Rabbi, Oluoch, Muranaka, Elzein, Odhiambo, Farombi and Gedil2017). Botanga et al. (Reference Botanga, Kling, Berner and Timko2002) reported low diversity among and between S. asiatica populations collected from across Benin, although there were strong correlations between geographic and genetic distances.
Striga gesnerioides races have been classified based upon their genetic relatedness and their capacity to differentially parasitize cowpea varieties and landraces (Botanga and Timko Reference Botanga and Timko2007; Lane et al. Reference Lane, Moore, Child, Cardwell, Singh and Bailey1993, Reference Lane, Moore, Child and Cardwell1996; Parker and Polniaszek Reference Parker and Polniaszek1990). Three races of the parasite were originally established based upon the differential responses of two Striga-susceptible and three Striga-resistant cowpea lines to the parasite (Parker and Polniaszek Reference Parker and Polniaszek1990), and a fourth race was reported shortly thereafter (Lane et al. Reference Lane, Moore, Child, Cardwell, Singh and Bailey1993). Examination of 48 Striga populations collected from 1984 to 1993 from seven countries in West Africa led Lane et al. (Reference Lane, Moore, Child and Cardwell1996) to propose the presence of at least five races (designated SG1 through SG5; Table 1) of the parasite based upon the differential responses of one Striga-susceptible and three Striga-resistant cowpea lines to parasitism. Races SG1 and SG5 were the most widespread, while SG4 was only detected in the Zakpota region of Benin (Lane et al. Reference Lane, Moore, Child and Cardwell1996). Striga gesnerioides race structure among 24 populations was reassessed by combining the results of differential parasitism of six resistant and three susceptible cowpea lines with molecular analysis of Striga genetic diversity, leading Botanga and Timko (Reference Botanga and Timko2007) to conclude that intra- and interpopulation variability was low in S. gesnerioides. Based on the nearly identical genotypes of populations within and outside of the Zakpota region of Benin, and the ability of these populations to differentially parasitize unique hosts, these researchers proposed SG4 as the designation of the race in the greater part of Benin and SG4z for the race from Zakpota (Table 1). In their analysis, Botanga and Timko (Reference Botanga and Timko2007) did not account for the fact that the newly defined SG4 population had parasitized the same cowpea varieties as the SG1 race previously defined by Lane et al. (Reference Lane, Moore, Child and Cardwell1996). They also suggested that Striga populations from Senegal be designated SG6 based primarily on genotypic characteristics and geographic location. However, the differential parasitism of variably resistant cowpea lines indicated SG6 corresponded to the SG1 designation by Lane et al. (Reference Lane, Moore, Child and Cardwell1996). Because differential parasitism of cowpea lines was only tested for Striga populations collected from Senegal by Botanga and Timko (Reference Botanga and Timko2007), it is unknown how the host compatibilities of the other genotyped Striga populations correspond to races designated by Lane et al. (Reference Lane, Moore, Child and Cardwell1996).
Table 1. Proposed and historical race designation for Striga gesnerioides in West Africa based on differential host parasitism.
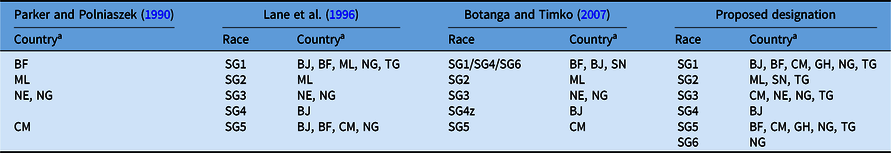
a Abbreviations: BF, Burkina Faso; BJ, Benin; CM, Cameroon; GH, Ghana; ML, Mali; NE, Niger; NG, Nigeria; SN, Senegal; TG, Togo.
Currently, there are limited sources of Striga resistance in cowpea germplasm. One of the most widely employed and best characterized Striga resistance genes in cowpea is RSG3-301, derived from the Botswanan landrace B301. B301 is resistant to most races of the parasite, with the exception of race SG4, to which it is susceptible (Botanga and Timko Reference Botanga and Timko2007; Lane et al. Reference Lane, Moore, Child and Cardwell1996; Li and Timko, Reference Li and Timko2009). Given the extensive use of this Striga resistance gene in cowpea breeding and the potential for resistance breakdown over time (as observed for B301 in Zakpota), a clear need exists to evaluate how the deployment of Striga-resistant cowpea varieties has impacted Striga diversity throughout West Africa and determine the effectiveness of known sources of Striga resistance. Furthermore, with the increased volume of grain exchanged between countries, the potential for parasite movement and expansion of races across West Africa need to be studied in order to provide clear and effective breeding targets. Thus, the current study was undertaken to better define current race structure for the parasite and to provide a more complete depiction of the relationship between genetic variation and virulence based on host specificity.
Materials and Methods
Host and Parasite Germplasm
Eight cowpea genotypes (524B, California Blackeye #5, 58-57, B301, IT82D-849, IT97K-499-35, IT81D-994, and Suvita-2) were used as hosts based on their reported resistance and susceptibility to different S. gesnerioides races (Botanga and Timko Reference Botanga and Timko2007; Lane et al. Reference Lane, Moore, Child, Cardwell, Singh and Bailey1993, Reference Lane, Moore, Child and Cardwell1996; Parker and Polniaszek Reference Parker and Polniaszek1990). Race designation in this study is based on the differential parasitism of cowpea varieties, breeding lines, and landraces with reported resistance to S. gesnerioides and corresponds to the previous designations defined by Lane et al. (Reference Lane, Moore, Child and Cardwell1996) (Table 1). 524B is a Striga-susceptible, blackeye variety developed at and acquired from the University of California, Riverside. California Blackeye #5 is a Striga-susceptible commercial variety and was purchased from Henry Field’s Seed & Nursery Company, Aurora, IN. The landrace 58-57 from Senegal is resistant to races SG4 and SG5. B301 is a landrace from Botswana that has been widely used as a source of Striga resistance by breeders and is only susceptible to S. gesnerioides race SG4 from the Zakpota region of Benin (designated SG4z by Botanga and Timko [Reference Botanga and Timko2007]). B301 was kindly provided by Lucky Omoigui at the University of Agriculture, Makurdi. A breeding line IT82D-849 and commercial variety IT97K-499-35, both developed by the International Institute of Tropical Agriculture (IITA) in Nigeria, are resistant to all races with the exception of SG4. The IITA breeding line, IT81D-994, and Burkinabe landrace, Suvita-2 (a.k.a. Gorom Local), are resistant to SG1, SG2, and SG4. Both lines were obtained from Benoit Batieno at the Institut de l’Environnement et de Recherches Agricoles.
All experiments involving viable S. gesnerioides seeds, developing parasites, and host–parasite interactions were performed in a USDA Animal and Plant Health Inspection Service (APHIS)-approved quarantine facility in the Department of Biology at the University of Virginia (Facility Number 669; APHIS Plant Protection and Quarantine Permit No. P526P-14-03013). Striga gesnerioides seeds were collected from nine countries in West Africa with the support of the West African Cowpea Consortium during the years 2005 to 2018 (Table 2). Bulked seed from at least 20 Striga stalks were collected from across each field site, thus providing a uniform representation of the diversity at each location and ensuring sufficient seed. Striga populations were selected based on their geographic location and seed viability to provide detailed coverage (Figure 1). In total, 58 Striga populations were tested against all cowpea lines, with the exception of California Blackeye #5, which was used as a susceptible host in a separate and accompanying experiment.
Table 2. Striga gesnerioides populations collected from 2005 to 2018 across nine West African countries.
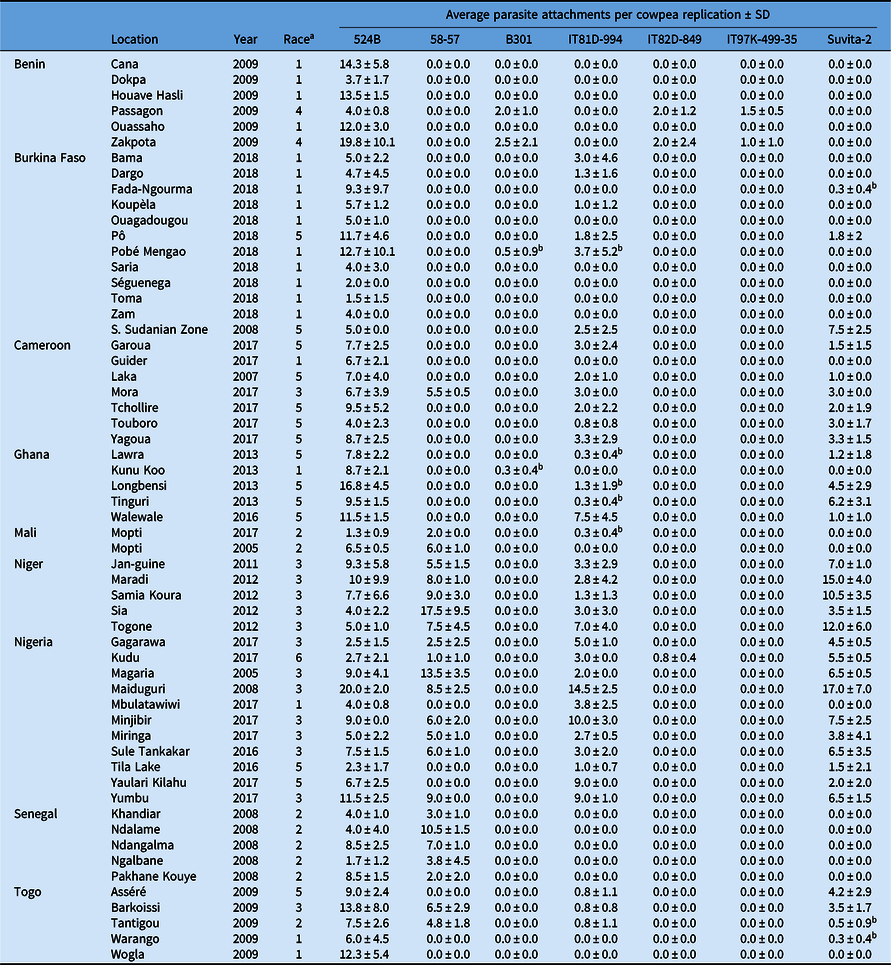
a Race was assigned based on the differential parasitism of seven cowpea lines.
b Attachments were only observed on a single replication and were unable to be replicated in subsequent trials.
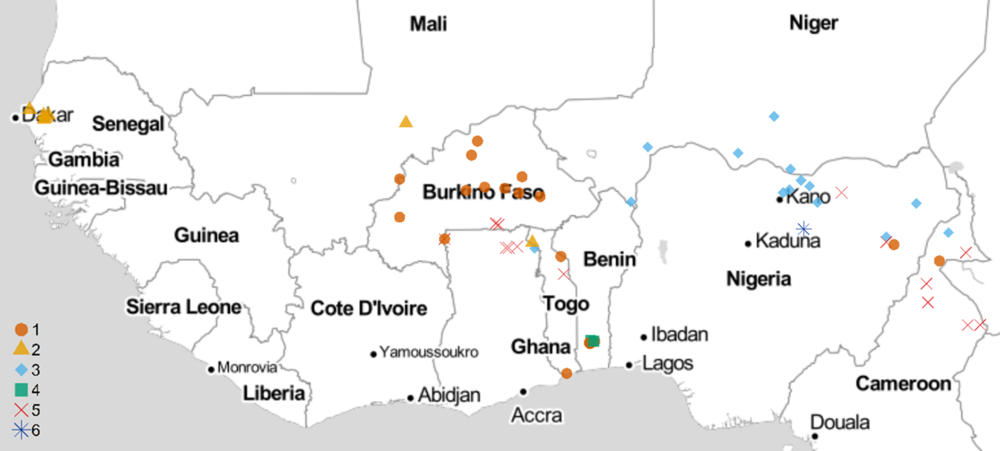
Figure 1. Striga gesnerioides sampling locations and race distribution across West Africa. The map was developed in R using the GGMAP package (Kahle and Wickham Reference Kahle and Wickham2013; R Core Team 2019) and stamen map tiles (map tiles by Stamen Design, under CC BY 3.0; data by OpenStreetMap, under ODbL)
Screening Experiments
D16H deepots (Stuewe & Sons, Tangent, OR) were filled with soil that consisted of approximately 70% sand and 30% metromix. Striga seeds were combined with fine sand (<250 microns) to approximate 2,000 seeds by weight per gram. One gram of the mixture was added to each pot. Pots were watered to maintain soil moisture and break Striga seed dormancy. All plants were grown on 12-h light:12-h dark photoperiods at 30 C.
After 7 d, two cowpea seeds were planted per pot in a replicated experiment for a total of four plants per cowpea accession. Each pot was considered a single replication, consisting of two plants. At 10 wk after cowpea planting, each pot was inspected for emerged Striga stalks. To access the presence of unemerged parasites, the cowpea plants were carefully uprooted and shaken free of soil to identify subterraneous Striga attachments. The total number of Striga attachments per replication was recorded based upon visual inspection of roots without magnification. For the purposes of these experiments, if attached parasites were observed across multiple replications, the cultivar was considered susceptible. However, if discrepancies between replications were observed, the experiment was repeated to ensure reproducibility. If parasites were observed only in a single replication, and if the susceptible phenotype was unable to be replicated in subsequent experiments, the cultivar was considered resistant. Suvita-2 was used as a proxy for IT81D-994 for race designation due to the detection of minimal heterogeneity among the IT81D-994 seed and inconsistency between replications for five of the Striga populations (Table 2). Suvita-2 contains the same Striga resistance locus as IT81D-994 (Ouédraogo et al. Reference Ouédraogo, Tignegre, Timko and Belzile2002b) and was used previously for race designation by Parker and Polniaszek (Reference Parker and Polniaszek1990).
Genetic Diversity Analysis
In a separate set of experiments, 10 Striga populations from seven countries were grown on California Blackeye #5 as described earlier, and individual stalks/leaves of emerged parasites were collected, flash frozen in liquid N2, and stored at −80 C for subsequent nucleic acid isolation. DNA was extracted from 5 to 13 emerged Striga plants from each population using a modified CTAB protocol (Doyle and Doyle Reference Doyle and Doyle1987). In total, DNA was collected from 87 unique Striga plants.
Twenty randomly distributed simple sequence repeat (SSR) markers (Supplementary Table S1) were developed from a preliminary draft genome of S. gesnerioides race SG3 using WebSat and default parameters (Martins et al. Reference Martins, Lucas, de Souza Neves and Bertioli2009). Each SSR corresponds to a unique scaffold to reduce the possibility of linkage between the markers and ensure random genomic distribution. In total, primers were developed for 18 perfect SSRs and two imperfect SSRs. Seven of the SSRs were trinucleotide repeats, while the remainder were dinucleotide repeats (Supplementary Table S1). For each of the 10 populations, DNA was combined in equal concentrations to develop pooled samples representative of each population. The primers were tested on each of the 10 pooled samples to ensure reproducible amplification and to optimize polymerase chain reaction (PCR) conditions. Those markers that consistently produced the same amplification patterns and that exhibited polymorphism among the 10 populations were selected for carrying out the rest of the experiment. In total, 11 polymorphic SSRs were deemed suitable for use in genotypic analysis based upon reproducible and strong amplification. The 87 DNA samples were genotyped with all 11 markers. The PCR amplifications were performed in 20-µl volumes consisting of 1 × Taq buffer, 200 µM dNTPs, 50 ng genomic DNA, 0.5 µM each primer, 2 mM MgCl2, and 1 unit Taq polymerase. Thermocycling consisted of 2-min denaturation (95 C), followed by 35 cycles of 45-s denaturation (95 C), 45-s annealing (60 C), and 45 = s extension (72 C), with a final extension time of 2 min (72 C). PCR amplicons were resolved by electrophoresis on polyacrylamide gels (80 g L−1) using a C.B.S. Scientific Mega-Gel System, stained with ethidium bromide, and visualized under ultraviolet light.
The presence/absence of amplicons was scored, and diversity analysis was performed using GenAlEx (Peakall and Smouse Reference Peakall and Smouse2006). Total and pairwise linear F ST, F′ST, Nei’s genetic distance/identity, Shannon’s diversity index, and principal coordinate analysis (PCoA) were used to characterize intra- and interpopulation diversity and population relatedness. Correlation between population diversity and the number of susceptible lines was tested for in Microsoft Excel 2016 (Microsoft, Redmond, WA).
Results and Discussion
Host Differential Response
Previous studies examined the race structure of S. gesnerioides populations in sub-Saharan West Africa using host differential-response assays (i.e., assessment of successful vs. unsuccessful parasitism) and molecular genotyping. These studies involving between 17 and 48 populations led to the identification of 4 to 7 distinct S. gesnerioides races based upon the differential parasitism of the Striga-resistant cowpea lines 58-57, B301, IT81D-994, and Suvita-2 (Botanga and Timko Reference Botanga and Timko2007; Lane et al. Reference Lane, Moore, Child and Cardwell1996; Parker and Polniaszek Reference Parker and Polniaszek1990) Given that race structure and distribution are dynamic and can be influenced by gene flow, genetic drift, and natural selection, we carried out a thorough investigation of S. gesnerioides diversity and virulence by screening seven cowpea lines against 58 unique S. gesnerioides populations collected across nine West African countries (Figure 1). We assigned the various parasite populations to races (pathotypes) based upon the host differential response to parasitism (Table 3). In the process, we attempted to ameliorate differences existing in prior literature regarding designations of races (e.g., between the works of Lane et al. [Reference Lane, Moore, Child and Cardwell1996] and Botanga and Timko [Reference Botanga and Timko2007]). Our findings indicate that there are likely a minimum of six races of S. gesnerioides based on the differential ability of various cowpea lines to support parasite attachment and growth (i.e., compatibility/incompatibility), with some races having genotypically distinguishable variants. The number of races and their geographic distribution indicate that despite a significant expansion in the volume of cowpea grain movement in the region (Langyintuo et al. Reference Langyintuo, Lowenberg-DeBoer, Faye, Lambert, Ibro, Moussa, Kergna, Kushwaha, Musa and Ntoukam2003), there has been little overall change in race distribution since the initial extensive surveys conducted more than two decades ago by Lane et al. (Reference Lane, Moore, Child and Cardwell1996).
Table 3. Resistance and susceptibility of seven cowpea lines to each of six defined Striga gesnerioides races.a

a Abbreviations: S, susceptible; R, resistant.
Race SG1 is the most widely distributed race across West Africa, occurring in six of the nine countries where collections were made, and is the primary race in Burkina Faso. Based on the results of host differential-response screenings, it is clear that the race designated SG4 by Botanga and Timko (Reference Botanga and Timko2007) is a genotypic variant of race SG1 and would be more appropriately designated as SG1-4 based on its genotypic similarity to race SG4 (Table 2). Race SG2 is present in Mali, Togo, and Senegal (formerly misclassified as SG6). Race SG3 predominantly occurs in Niger and northern Nigeria, and SG5 occurs in Cameroon and Ghana. SG3 and SG5 are present in at least four countries, and SG2 in three. Race SG4 is only detected near the Zakpota region of Benin. However, all six available populations from Benin were collected from the Zou Department of Benin and may not provide a complete picture of race distribution within the country. Race SG4 (formerly referred to as SG4z by Botanga and Timko [Reference Botanga and Timko2007]) is genotypically very similar to SG1-4, suggesting SG1-4 and SG4 share a recent and common ancestry, but are clearly distinguishable by the ability of race SG4 to overcome Striga resistance conferred by B301 (Tables 2, 4, and 5).
Table 4. F′ ST and F ST values of Striga gesnerioides populations from West Africa based on simple sequence repeat (SSR) markers.a
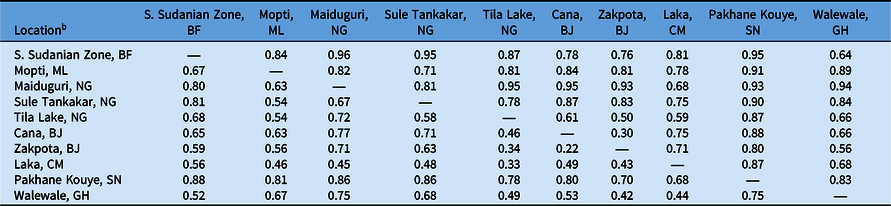
a F′ ST is reported above the diagonal; F ST is reported below the diagonal.
b Abbreviations: BF, Burkina Faso; BJ, Benin; CM, Cameroon; GH, Ghana; ML, Mali; NG, Nigeria; SN, Senegal.
Table 5. Pairwise comparisons of Nei’s genetic distance and Nei’s genetic identity of select Striga gesnerioides populations based on simple sequence repeat (SSR) markers.a
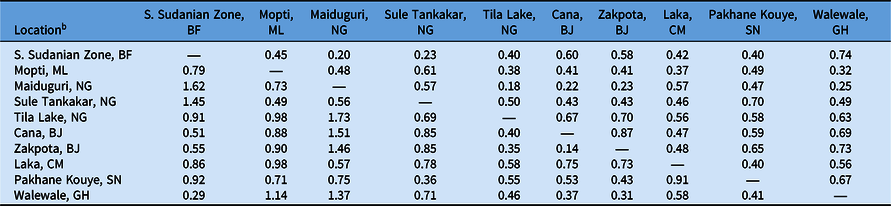
a Nei’s genetic distance is reported below the diagonal; Nei’s genetic identity is reported above the diagonal.
b Abbreviations: BF, Burkina Faso; BJ, Benin; CM, Cameroon; GH, Ghana; ML, Mali; NG, Nigeria; SN, Senegal.
In contrast to the results of prior studies, multiple races were detected in both Cameroon and Togo, where previously only single races were reported (e.g., SG5 in Cameroon and SG1 in Togo, according to Lane et al. [Reference Lane, Moore, Child and Cardwell1996]). The expansion in the number of races detected could be due to differences in the sampling locations or could result from the introduction or evolution of new Striga races over time. We also found two races, SG1 and SG5, in Ghana. Our observation of the existence of multiple races in Ghana is consistent with a recent examination of Striga diversity by Awuku (Reference Awuku2017), who reported the presence of races SG2, SG3, and SG5 among just eight parasite populations. While we did not detect SG2 or SG3 in Ghana, their presence is unsurprising, given both races were detected in neighboring Togo (Table 1; Figure 1). The greatest Striga diversity was detected in Togo, which contains at least four different races distributed throughout the country based on host response. Four races were found in Nigeria, three in Cameroon, and two each in Benin, Burkina Faso, and Ghana. The presence of a single race was detected in Mali (SG2), Niger (SG3), and Senegal (SG2), although in the case of Mali we had few available populations (Table 2; Figure 1). The existence of multiple races within the majority of cowpea-producing countries in West Africa emphasizes the importance of stacking multiple resistance genes to ensure durable Striga resistance.
A new hypervirulent race of the parasite that parasitized the highest number of cowpea lines (524B, 58-57, IT81D-994, IT82D-849, and Suvita-2) was collected from Kudu, Nigeria. This race (designated SG6) was found to parasitize five of the seven cowpea lines. Race SG6 overcomes the Striga resistance of IT82D-849, which was previously only overcome by race SG4. Surprisingly, B301 and IT97K-499-35 were resistant, despite correspondence of resistance or susceptibility for all other populations between the three lines (Tables 2 and 3). This is especially unexpected based upon evidence supporting the presence of the RSG3-301 Striga resistance gene in each of B301, IT97K-499-35, and IT82D-849. While previously it was reported that resistance in IT82D-849 was non-allelic to B301 (Atokple et al. Reference Atokple, Singh and Emechebe1995), most other evidence suggests the Striga resistance locus of IT82D-849 and B301 coincide or are tightly linked. Genotyping of IT82D-849 with SSR1, a marker that is reportedly specific to the B301 RSG3-301 allele (Li and Timko Reference Li and Timko2009), indicates the presence of the allele in IT82D-849 (unpublished data). Additionally, the SG1 resistance gene has been mapped to the same genomic locus in IT82D-849 as in B301 (Ouédraogo et al. Reference Ouédraogo, Maheshwari, Berner, St-Pierre, Belzile and Timko2001). As the source of Striga resistance in IT82D-849 is reportedly Emma 60, not B301, IT82D-849 may contain an alternative RSG3-301 allele or B301 and IT97K-499-35 may contain additional resistance genes or quantitative trait loci that modify the levels of Striga resistance (Atokple et al. Reference Atokple, Singh and Emechebe1995). Alternatively, B301 and IT97K-499-35 may have escaped susceptibility to the Kudu population based upon the low average number of individuals (0.8 ± 0.4) that parasitized IT82D-849 (Table 2). Despite our replication of the Kudu screening experiment a second time, no Striga attachments were observed on any of the four B301 or IT97K-499-35 replications, although parasitism was detected on three of the four IT82D-849 replications (Table 2).
It is unclear whether the Striga populations used in this and previous studies compose distinct races or are combinations of multiple races. Due to the difficulty of procuring and developing pure Striga lines by single-seed descent, the populations used in this and previous studies were bulks of seeds collected from multiple plants within a local population. For instance, the SG2 race combined with SG5 could result in classification as race SG3 (Table 3). Of the 46 instances in which the Striga populations were compatible with more than one cultivar, the average number of supported attachments was just 64% of that found for the susceptible control. Combinations of multiple races may explain the greater numbers of Striga plants supported by 524B on average compared with cowpea varieties containing at least one resistance gene. Striga resistance genes could also confer partial resistance, leading to fewer attachments. Development of inbred Striga lines would resolve this uncertainty.
Effectiveness of Cowpea Striga Resistance Genes
As expected from prior studies, 524B was susceptible to all Striga populations. None of the cowpea lines tested in this study are resistant to all 58 Striga populations (Table 2; Figure 1). Varieties 58-57, IT81D-994, and Suvita-2 were each susceptible to approximately half of the populations, although they complement more broadly resistant cultivars that are susceptible to SG4. In most cases, Suvita-2 and IT81D-994 were resistant or susceptible to the same populations. However, seed of IT81D-994 did not appear to be completely homogeneous based upon inconsistency between replications for five populations (Table 2). Genotyping of several IT81D-994 individuals using the Cowpea iSelect Consortium Array confirmed that there was genomic heterozygosity present (unpublished data), and we therefore used Suvita-2 as a proxy for IT81D-994 for race designation. Suvita-2 has been previously reported to be a suitable proxy for IT81D-994 (Parker and Polniaszek Reference Parker and Polniaszek1990). Landrace 58-57 presents a unique resistance fingerprint and likely contains unique resistance genes/alleles. IT81D-994 and Suvita-2 are susceptible to SG3, SG5, and SG6, while 58-57 is susceptible to SG2, SG3, and SG6. Striga gesnerioides resistance genes from Suvita-2 and IT81D-994 were mapped to chromosome Vu10, while the genomic location of the Striga resistance in 58-57 is unknown (Ouédraogo et al. Reference Ouédraogo, Maheshwari, Berner, St-Pierre, Belzile and Timko2001, Reference Ouédraogo, Gowda, Jean, Close, Ehlers, Hall, Gillaspie, Roberts, Ismail, Bruening, Gepts, Timko and Belzile2002a, Reference Ouédraogo, Tignegre, Timko and Belzile2002b). B301, IT97K-499-35, and IT82D-849 confer the broadest spectrum of resistance and exhibit similar resistance fingerprints, with the exception of the susceptibility of IT82D-849 to SG6 (Table 3). The B301, IT82D-849, and IT97K-499-35 Striga resistance genes colocalize on chromosome Vu11 (Asare et al. Reference Asare, Galyuon, Padi, Otwe and Takrama2013; Li and Timko Reference Li and Timko2009; Ouédraogo et al. Reference Ouédraogo, Maheshwari, Berner, St-Pierre, Belzile and Timko2001). Because none of the reported races of Striga are capable of overcoming all known sources of Striga resistance in cowpea, complete and durable resistance can be achieved by stacking multiple resistance genes.
Genetic Diversity Analysis
To determine the extent of intra- and interpopulation diversity among the various Striga populations collected in this study, we measured the genetic diversity present within several Striga populations using SSRs. Our analyses indicate that both intra- and interpopulation diversity existed and were easily detected within and between all 10 populations using as few as five individuals per population and 11 SSR markers (Tables 4–6). As expected in a primarily autogamous species, significant genetic differentiation exists among S. gesnerioides populations across West Africa, as indicated by F ST of 0.633 and F′ ST of 0.810. Pairwise linear F ST and F′ ST indicate that the only exceptions to the high level of differentiation between populations are the populations from Benin, which exhibit only moderate differentiation. This is likely a reflection of the proximity of collection sites (Table 4). The similarity of the Benin populations is also supported by a Nei’s genetic identity of 0.873 (Table 5). Highly similar populations were also detected in comparisons of Striga from Walewale (Ghana) with populations from the South Sudanian Zone of Burkina Faso and Zakpota (Benin), and comparisons of Tila Lake (Nigeria) with Zakpota and Walewale.
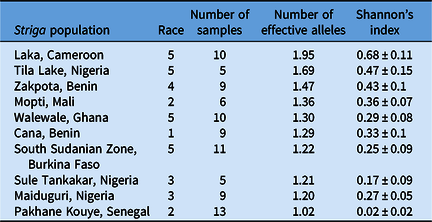
Table 6. Diversity within select Striga gesnerioides populations from West Africa based on simple sequence repeat (SSR) genotyping.
While in some instances genetic identity corresponds with geographic distance, this is not always the case. Two Nigerian populations, Tila Lake and Maiduguri, are highly dissimilar, sharing a genetic identity of just 0.177, and belong to different races despite their close geographic proximity. The seeds from these two locations were collected 8 yr apart, suggesting temporal distance may also have contributed to lower genetic similarity between these two populations. High dissimilarity was also detected when comparing Burkina Faso with Maiduguri and Sule Tankakar and comparing Maiduguri with Cana (Benin), Zakpota, and Walewale (Table 5). The high genetic dissimilarity between these other populations is unsurprising, given the large distances between sampling locations and their varied race assignments.
To determine whether genetic differentiation among populations corresponded with race, we compared the genetic relatedness of populations with their differential parasitism of seven cowpea lines. The results of this study indicate that the genetic similarity of Striga populations based on randomly distributed SSR markers is unsuitable for race assignment. While the populations from Cana and Zakpota in Benin are genetically very similar, they belong to different races. Correspondingly, populations from Pakhane Kouye (Senegal) and Mopti (Mali) each are race SG2, but only share moderate genetic relatedness, likely in part due to the distance and time between samplings (Figure 1; Table 5). While populations from Burkina Faso, Nigeria, Cameroon, and Ghana are all race SG5, the genetic identity among them varies considerably and their similarity resembles the average genetic identity across all populations (Table 5).
The most genetically diverse genotyped population was from Laka, Cameroon, while Pakhane Kouye, Senegal, was the least diverse based on Shannon’s diversity index and PCoA (Table 6; Figure 2). However, Shannon’s diversity could be skewed somewhat by sampling methodology, as this was not entirely standardized across populations. The number of Striga plants used to establish these experimental populations and the cowpea hosts they were collected from likely influenced the levels of diversity. Diversity ranged significantly from 0.02 to 0.68 based on Shannon’s diversity index and the number of effective alleles (Ne) ranged from 1.02 to 1.95 (Table 6). However, there was little correlation between population diversity and the number of compatible host lines (r = 0.01).
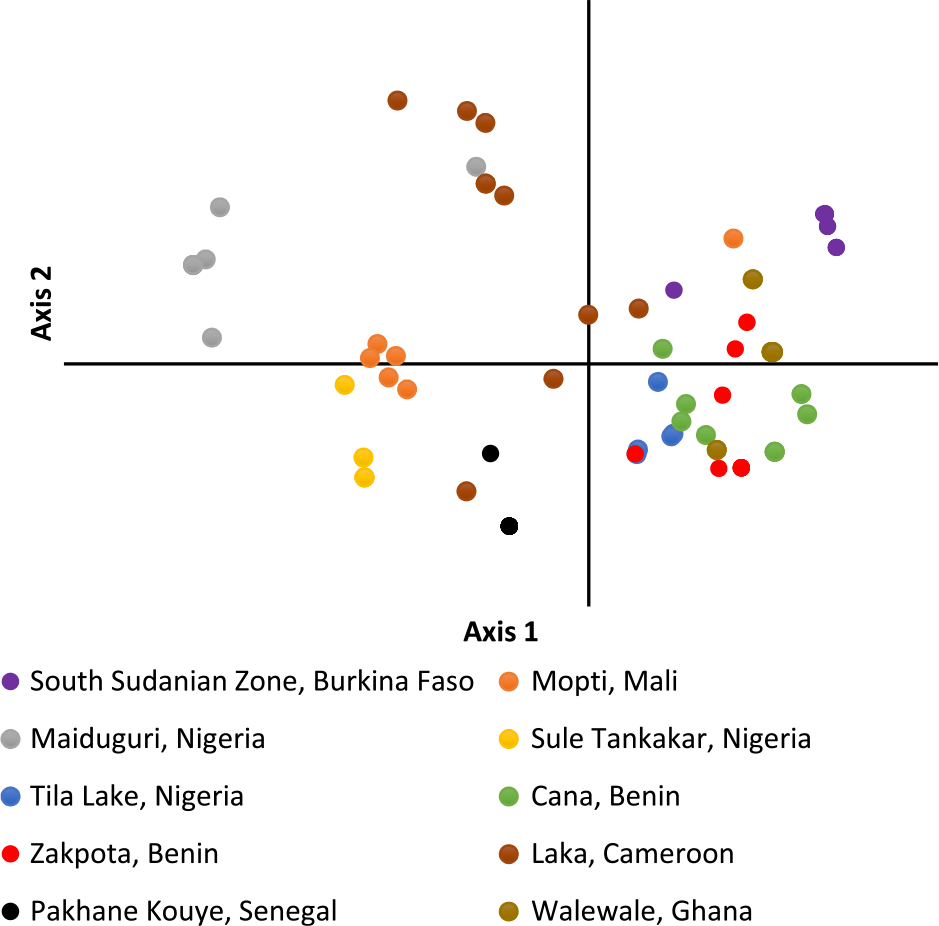
Figure 2. Principal coordinate analysis of individual Striga gesnerioides samples from 10 populations.
PCoA of individual Striga plants based on the first two axes indicates samples within the same population cluster together, with the exception of a population from Cameroon (Figure 2). The first axis explains 22.9% of the variation, while the second axis explains 17.6%. Population-level PCoA suggests the populations from Tila Lake (Nigeria), Cana (Benin), Zakpota (Benin), and Walewale (Ghana) are highly similar (Figure 3). Axis 1 explains 34.7% of the variation, and axis 2 explains 19.6%. In agreement with Nei’s estimates of genetic relatedness, PCoA indicates little association between race and genetic relatedness.
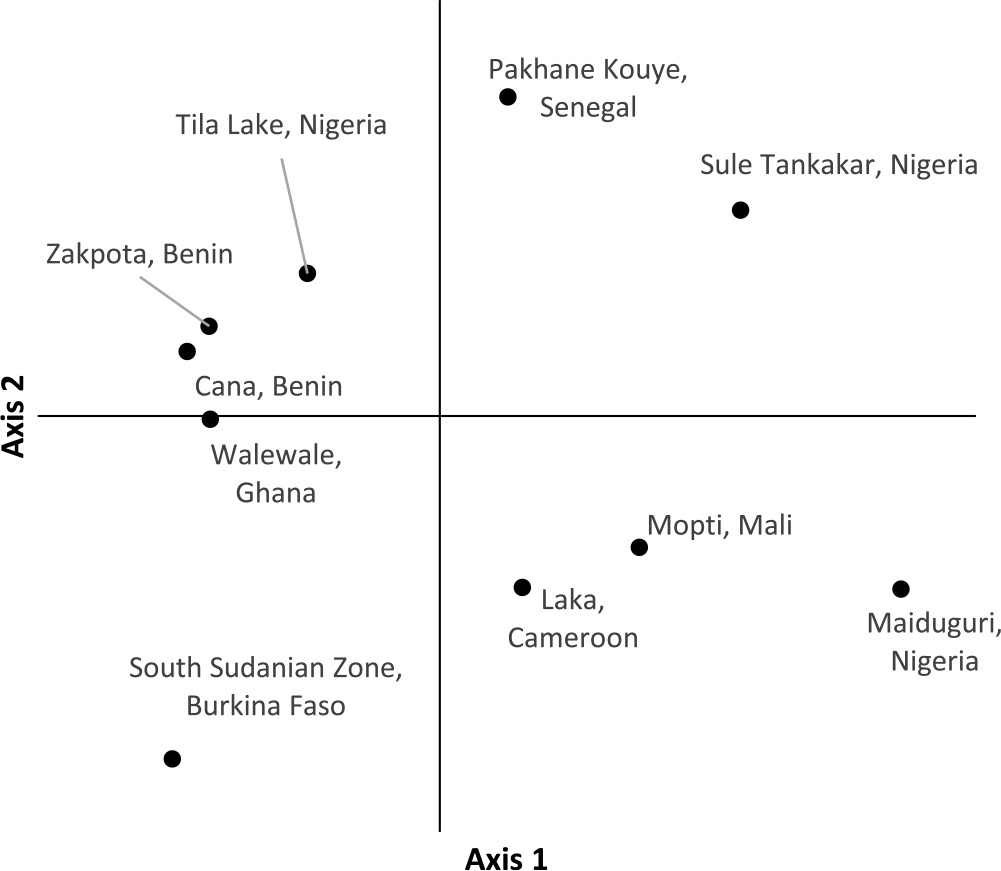
Figure 3. Principal coordinate analysis of 10 Striga gesnerioides populations from West Africa.
Based on the differential parasitism of seven cowpea lines, at least six unique Striga races are present in West Africa. Although populations are distinguishable genetically from one another using molecular markers, including AFLPs, SSRs, and single-nucleotide polymorphisms, genetic relatedness does not generally correspond with host specificity and appears more indicative of geographic origin or date of collection (Bharathalakshmi et al. 1990; Unachukwu et al. Reference Unachukwu, Menkir, Rabbi, Oluoch, Muranaka, Elzein, Odhiambo, Farombi and Gedil2017; Welsh and Mohamed Reference Welsh and Mohamed2011). From a breeding perspective, the wide distribution of S. gesnerioides races indicates the need to stack multiple Striga resistance genes to ensure durable resistance throughout West Africa. At least three and potentially four unique resistance genes or alleles have been identified among the seven cowpea lines examined in this study based on their unique resistance fingerprints (Table 3). Although none of the cowpea lines are resistant to all Striga races, even the most virulent races are unable to overcome all known sources of Striga resistance in cowpea. Fortunately, the predominant race, SG1, is unable to overcome any of the known sources of Striga resistance in cowpea. However, we already see evidence of resistance breakdown among the hypervirulent races SG4 and SG6 from Benin and Nigeria, respectively. Fortunately, the only Striga populations capable of overcoming the B301 source of resistance were confined to a single region of Benin. Continued monitoring of Striga in West Africa and identification and deployment of new sources of resistance in cowpea are needed.
Supplementary material
To view supplementary material for this article, please visit https://doi.org/10.1017/wsc.2020.3
Acknowledgments
The authors wish to thank Philip Roberts (University of California, Riverside), Lucky Omoigui (University of Agriculture, Makurdi), and Benoit Joseph Batieno (Institut de l’Environnement et Recherches Agricoles) for providing the cowpea germplasm used in this study. We also wish to acknowledge the members of the West African Cowpea Consortium, especially Lucky Omoigui, Sobda Gonne, Sory Diallo, Benoit Joseph Batieno, Moctar Wade, Frank Essem, Nana Aicha Coulibaly, and Béré Tchabana, who provided valuable collections of parasite seeds for this analysis over the past decade, and the members of the Kirkhouse Trust Scottish Charitable Incorporated Organisation (SCIO), who supported these efforts. Many thanks also to Matthew Tolerico for assisting with setting up experiments and Eli Connell for assisting with tissue collection, DNA extraction, and SSR genotyping. This research was supported by a grant from the Kirkhouse Trust SCIO to MPT. MPT was also supported in part by funds from the National Science Foundation Plant Genome Research Program (IOS-1238057). No conflicts of interest have been declared.











