1. Introduction
Transposable elements are present in the genomes of many organisms, including model genetic species such as Drosophila melanogaster. The population of transposons in this organism comprises elements that move through the agency of a transposase enzyme (the cut-and-paste transposons) and elements that move by producing DNA copies of their RNA transcripts (the retrotransposons). Among the hundred or so different types of transposons in the Drosophila genome, only a few have been intensively studied. One of them, the cut-and-paste transposon called the P element, has become a valuable tool for genetic analysis in Drosophila. It has also provided important insights into how Drosophila transposons are regulated.
P elements were discovered through their involvement in the phenomenon of hybrid dysgenesis, a syndrome of abnormal germ-line traits that occurs non-reciprocally in the offspring of crosses between certain types of Drosophila strains (Kidwell et al., Reference Kidwell, Kidwell and Sved1977; Engels, Reference Engels, Berg and Howe1989). P strains contain P elements in their genomes, whereas M strains do not. Crosses between P males and M females produce dysgenic offspring, whereas crosses between M males and P females and between P males and P females generally do not. The traits of hybrid dysgenesis – agametic sterility, increased frequencies of mutation and chromosome breakage, transmission ratio distortion and chromosome non-disjunction – occur in the hybrid offspring of P males and M females because the P elements that are contributed by the males in these crosses are not repressed in the offspring. In the other crosses, a maternally contributed state called the P cytotype represses P-element activity in the offspring (Engels, Reference Engels1979).
Recent advances in genomic analysis have implicated small RNAs in the repression of transposon activity (Brennecke et al., Reference Brennecke, Aravin, Stark, Dus, Kellis, Sachidanandam and Hannon2007, Reference Brennecke, Malone, Aravin, Sachidanandam, Stark and Hannon2008; Li et al., Reference Li, Vagin, Lee, Xu, Ma, Xi, Seitz, Horwich, Syrzycka, Honda, Kittler, Zapp, Klattenhoff, Schulz, Theurkauf, Weng and Zamore2009). These RNAs associate with the Piwi class of proteins, which in Drosophila includes the denominative Piwi and two other members, Aubergine and Argonaute3. These RNAs are therefore called Piwi-interacting or piRNAs. Certain loci in the Drosophila genome produce piRNAs. One especially productive locus is situated in the telomere-associated sequences (TAS) at the end of the left arm of the X chromosome (XL). When P elements are inserted in this locus, they participate in the generation of piRNAs. Because the P-specific piRNAs derived from this and other loci are transmitted maternally through the egg cytoplasm (Brennecke et al., Reference Brennecke, Malone, Aravin, Sachidanandam, Stark and Hannon2008), they are thought to be the physical basis of the P cytotype (Jensen et al., Reference Jensen, Stuart, Goodpaster, Goodman and Simmons2008). In the offspring of crosses involving P females, antisense piRNAs may repress hybrid dysgenesis by targeting P-element mRNAs, including those that encode the P transposase, for destruction. P-specific piRNAs may also guide protein complexes to P elements inserted at diverse locations in the genome to prevent their movement.
The genetic analysis of telomeric P elements has revealed the important role they play in P-element regulation (Ronsseray et al., Reference Ronsseray, Lehmann and Anxolabéhère1991, Reference Ronsseray, Lemaitre and Coen1993, Reference Ronsseray, Lehmann, Nouaud and Anxolabéhère1996, Reference Ronsseray, Marin, Lehmann and Anxolabéhère1998; Marin et al., Reference Marin, Lehmann, Nouaud, Izaabel, Anxolabéhère and Ronsseray2000; Stuart et al., Reference Stuart, Haley, Swedzinski, Lockner, Kocian and Simmons2002; Simmons et al., Reference Simmons, Raymond, Niemi, Stuart and Merriman2004; Niemi et al., Reference Niemi, Raymond, Patrek and Simmons2004; Thorp et al., Reference Thorp, Chapman and Simmons2009; Belinco et al., Reference Belinco, DiPrima, Wolff, Thorp, Buschette and Simmons2009). We have endeavoured to extend this analysis by examining the effects of mutations in the genes aubergine (aub), piwi and Suppressor of variegation 205 [Su(var)205] on the maternal component of this regulation. The genes aub and piwi encode Piwi-type proteins that associate with piRNAs (Brennecke et al., Reference Brennecke, Aravin, Stark, Dus, Kellis, Sachidanandam and Hannon2007); Su(var)205 encodes a protein called heterochromatin protein 1 (HP1), which is involved in chromatin organization (James et al., Reference James, Eissenberg, Craig, Diethrich, Hobson and Elgin1989). Heterozygous mutations in all three genes have been shown to impair the repression of hybrid dysgenesis (Ronsseray et al., Reference Ronsseray, Lehmann, Nouaud and Anxolabéhère1996; Reiss et al., Reference Reiss, Josse, Anxolabéhère and Ronsseray2004; Haley et al., Reference Haley, Stuart, Raymond, Niemi and Simmons2005; Simmons et al., Reference Simmons, Ryzek, Lamour, Goodman, Kummer and Merriman2007; Belinco et al., Reference Belinco, DiPrima, Wolff, Thorp, Buschette and Simmons2009).
In this paper, we address how the aub, piwi and Su(var)205 mutations impair P-element regulation. First, by monitoring excisions of particular P elements in the germ line, we document that this regulation depends on a maternal component. Second, using the excision assay, we investigate the maternal effects of aub, piwi and Su(var)205 mutations on P regulation. Finally, to elucidate the underlying molecular mechanism, we examine the effects of these mutations on the expression levels of specific P-element mRNAs in the female germ line.
2. Materials and methods
(i) Drosophila stocks and husbandry
Information on the special chromosomes and mutant alleles used in the experiments is available on the Flybase website, in Lindsley & Zimm (Reference Lindsley and Zimm1992), or in other references cited in the text. Experimental cultures were reared in vials on a standard cornmeal–molasses–dried yeast medium; the culturing temperatures are specified in the text.
TP5 is a 1·8-kb-long P element inserted in the TAS at the left end of an X chromosome marked with the tightly linked recessive mutation w (white eyes); see Stuart et al. (Reference Stuart, Haley, Swedzinski, Lockner, Kocian and Simmons2002) for the isolation and characterization of this telomeric P element. The TP5 w X chromosome was incorporated into a Gla/CyO stock, and later, into stocks carrying aub, piwi or Su(var)205 mutations balanced with the CyO chromosome II (Belinco et al., Reference Belinco, DiPrima, Wolff, Thorp, Buschette and Simmons2009). Control stocks were subsequently established by removing the mutations from the genotype (Belinco et al., Reference Belinco, DiPrima, Wolff, Thorp, Buschette and Simmons2009). A TP5 w snw; Gla/CyO stock was established by exchanging the CyO balancer for the Cy Roi balancer in a TP5 w snw; Gla/Cy Roi stock (Simmons et al., Reference Simmons, Ryzek, Lamour, Goodman, Kummer and Merriman2007) and a control w snw; Gla/CyO stock was established in the same way. In all these stocks, the CyO chromosome carried P(SB)7, an insertion of a P transgene that contains the Drosophila mini-white gene sandwiched between the terminal inverted repeats of a vertebrate transposon called Sleeping Beauty (Ivics et al., Reference Ivics, Hackett, Plasterk and Izsvák1997). This insertion, abbreviated SB7, confers a dark orange eye colour on flies in which the native white gene is mutant, and it becomes mobile when a source of the P transposase is present in the genome.
P(ry +, ▵2-3)99B, a P transgene abbreviated ▵2-3, is a stable source of the P transposase inserted on chromosome III (Robertson et al., Reference Robertson, Preston, Phillis, Johnson-Schlitz, Benz and Engels1988). H(hsp/CP)2, a hobo transgene abbreviated CP, is another stable source of the P transposase inserted in chromosome II (Simmons et al., Reference Simmons, Haley, Grimes, Raymond and Niemi2002 a). This transgene produces the P transposase only in the germ line. ▵2-3, by contrast, produces it in both the soma and the germ line. The somatic activity of ▵2-3 can make it difficult to score phenotypes that reveal P-element activity. Consequently, when ▵2-3 was used to induce P activity, these phenotypes were scored in genotypes that repress somatic transposase activity (Robertson & Engels, Reference Robertson and Engels1989).
(ii) P-element excision assay
The singed-weak (snw) allele is a double P-element insertion mutation of the X-linked singed bristle (sn) gene (Roiha et al., Reference Roiha, Rubin and O'Hare1988). This allele causes a mild malformation of the bristles on the adult cuticle. In the presence of the P transposase, either of the two P elements inserted in snw can be excised. Excision of one element produces a more extreme mutant phenotype (sne), whereas excision of the other element produces a pseudo-wild phenotype (sn(+)). Germ-line P-excisions from snw were detected by crossing an snw male to females with attached X chromosomes. The male's sons, which inherited their X chromosome patroclinously, were then scored for the three possible bristle phenotypes. The proportion of sn(+) and sne flies among these sons (a statistic referred to as the ‘snw mutability’) was used to quantify the male's germ-line P-excision activity.
(iii) P-transgene excision assay
Flies with the CyO, SB7 chromosome and a mutant native w gene have curly wings and coloured eyes. If the SB7 transgene is excised from the CyO chromosome in a male's germ line, some of the male's curly-winged offspring will have white eyes. The proportion of white-eyed curly flies among all the curly offspring can therefore be used as a measure of P-transgene excision activity in the male's germ line. This statistic will, of course, underestimate the true excision activity because some of the excised SB7 transgenes may re-integrate, either on the CyO chromosome or on another chromosome, and then be passed on to the offspring, which will develop coloured eyes. No attempt was made to analyse such reintegration events in the experiments reported here.
(iv) Statistical analyses
Excision frequencies for the snw P elements and the SB7 transgene were calculated independently for each replicate culture in an experimental group and the unweighted average frequency among the replicates was used to characterize that group. The standard error (se) for this average was computed empirically from the variance among replicates. Statistical differences between groups were assessed by performing t or z tests.
(v) RNA isolation and reverse transcription (RT)–PCR
RNA was isolated from groups of 20 virgin females using TRIZOL (Invitrogen) according to the supplier's instructions. The RNA was reverse transcribed into cDNA using the ThermoScript reverse transcriptase (Invitrogen) and an oligo-dT primer, and the resulting cDNA was amplified by the PCR over 30 cycles using appropriate oligonucleotide primers and temperature profiles. The detailed methods for RT–PCR and the P-element primer sequences are given in Jensen et al. (Reference Jensen, Stuart, Goodpaster, Goodman and Simmons2008) . The sequences of the primers that were used to amplify singed cDNA are: sn-u 5′-CGTATCTCCTTGGGTCTATCAACG-3′ and sn-d 5′-CTGGTCATCTGTTTGCCACCTC-3′. These primers anneal to segments of different exons in the singed gene. The PCR profile for amplification of singed cDNA was 3 min at 92°C, 2 min at 60°C and 3 min at 72°C followed by 29 cycles consisting of 1 min at 92°C, 2 min at 60°C and 3 min at 72°C. All PCR products were analysed in 1% agarose gels by electrophoresis at 70 volts.
3. Results
(i) A maternal component in the repression of P-element and P-transgene excisions in males
The capacity for P-element regulation in the male germ line has previously been studied by monitoring the frequency of transposase-induced P-element excisions from snw, a double P-insertion allele of the X-linked singed locus. These studies have indicated that regulation occurs when the males inherit an X-linked telomeric P-element maternally; however, when they inherit the telomeric P-element paternally, regulation is lost (Simmons et al., Reference Simmons, Raymond, Niemi, Stuart and Merriman2004). As a basis for further genetic and molecular analysis of P-element regulation, we confirmed the requirement for maternal inheritance of the telomeric P-element by simultaneously monitoring excisions of the P elements from snw and excisions of the P transgene, SB7, from the CyO balancer chromosome II. These excisions were induced in males by the ▵2-3 transposase source inserted in chromosome III, and regulatory capacity was provided by the telomeric P element TP5. The results of this two-locus assay for repression of P excisions are summarized in Table 1.
Table 1. Germ-line P-element and P-transgene excision in males carrying a maternally or a paternally inherited telomeric TP5 element

The (TP5) w snw; CyO, SB7/+; ▵2-3/+ males for these tests were obtained from “reciprocal” crosses between (TP5) w snw; CyO, SB7/Gla and w; ▵2-3 strains at 18°C: (TP5) w snw; CyO, SB7/Gla females x w; ▵2-3 males (maternal transmission) and (TP5) w snw; CyO, SB7/Gla males×C(1)DX, y w f; ▵2-3 females (paternal transmission). Each tested male was crossed to four C(1)DX, y f females from a P strain at 25°C. After 5 days the flies were transferred to a fresh culture. The sons of both cultures were scored for their bristle and eye colour phenotypes on days 14 and 17 after the cultures were established. Summary statistics were calculated after pooling the data from the two cultures.
a Unweighted average frequency of phenotypically sn + and sne sons among all sons scored ±se.
b Unweighted average frequency of white-eyed sons among curly-winged sons scored ±se.
In control tests, in which TP5 was absent, the frequency of P-element excisions, measured by the mutability of snw, was around 0·5, and the frequency of P-transgene excisions, measured by the loss of the SB7 transgene from the CyO chromosome, was around 0·3, regardless of the parental origin of the transposase targets (snw and SB7) or the transposase source (▵2-3). In males that had inherited TP5 maternally, these excision statistics were dramatically reduced (snw mutability=0·11 and SB7 excision frequency=0·06), but in males that had inherited TP5 paternally, they were essentially the same as those from males that did not carry TP5. The difference between the two types of TP5-bearing males cannot be attributed to cytoplasmic transmission of P transposase activity in the cross where ▵2-3 was inherited maternally and TP5 was inherited paternally because no such transmission is seen with the ▵2-3 transposase source (Simmons et al., Reference Simmons, Haley and Thompson2002b). Repression of both P-element and P-transgene excisions in the male germ line therefore requires the maternal inheritance of the TP5 element. Other studies employing the snw mutability assay have shown that repression of P excisions cannot be explained by a simple maternal effect of TP5 (Stuart et al., Reference Stuart, Haley, Swedzinski, Lockner, Kocian and Simmons2002; Thorp et al., Reference Thorp, Chapman and Simmons2009); rather, it requires the combined maternal and zygotic effects of this telomeric P element.
It should also be noted that all the males tested in this assay exhibited pronounced mosaicism for the bristle and eye colour phenotypes, even when they inherited TP5 maternally. This observation confirms published evidence that TP5 does not repress P excisions induced by the transposase that ▵2-3 produces in somatic tissues (Stuart et al., Reference Stuart, Haley, Swedzinski, Lockner, Kocian and Simmons2002; Simmons et al., Reference Simmons, Raymond, Niemi, Stuart and Merriman2004). In the sons of these males, however, the somatic mosaicism was suppressed by the P genetic background inherited from their mothers.
(ii) Maternal effects of mutations on repression of P excisions in males
We used the P-transgene excision assay to determine if the maternal component of P-element regulation requires the proteins encoded by the genes aub, piwi and Su(var)205, which are all located in chromosome II. We monitored excisions of the SB7 transgene in the sons of TP5 w females that were heterozygous for the CyO, SB7 chromosome II and a mutation in one of these genes. These TP5 w; CyO, SB7/mutation females were crossed to w males homozygous for a source of the P transposase to obtain the TP5 w; CyO, SB7/+ males that were tested. SB7 excisions in the germ lines of these males should be repressed by the zygotic and maternal effects of the maternally inherited TP5 element. However, if the heterozygous mutation present in the mother of each tested male depletes a protein that is essential for the maternal effect, the repression mechanism would be impaired and the SB7 excision frequency would increase.
We tested two aub mutations, two piwi mutations and one Su(var)205 mutation in two different experiments, one using the ▵2-3 transposase source (Table 2) and the other using the CP transposase source (Table 3). As a control, we used a stock carrying the Gla mutation, which has not been implicated in any aspect of transposon regulation. In both experiments, we also tested stocks that did not carry the TP5 element to determine if the various mutations influenced transgene excision frequency in the absence of P-element regulation.
Table 2. Maternal effects of heterozygous mutations on TP5-mediated repression of ▵2-3-induced P-transgene excision in the male germ line

Mutant stocks with and without the TP5 element were used in this experiment. The tested (TP5) w; CyO, SB7/+; ▵2-3/+ males were obtained by crossing (TP5) w; CyO, SB7/mutation females with w; ▵2-3 males at 18°C. Each tested male was crossed to three females from the Harwich w P strain at 25°C and the curly-winged offspring were scored for their eye colour phenotype on day 14. Although all the tested males were mosaic for eye colour, their offspring, which inherited the P genetic background from their mothers, were not. Asterisks indicate excision frequencies that are significantly greater than the frequency for the Gla mutation at the 5% level.
a Unweighted average frequency of white-eyed flies among curly-winged flies ±se.
Table 3. Maternal effects of heterozygous mutations on TP5-mediated repression of CP-induced P-transgene excision in the male germ line

The tested (TP5) w; CyO, SB7/CP males were obtained by crossing (TP5) w; CyO, SB7/(mutation) females with w; CP males at 25°C. Each tested male was crossed to three Harwich w females at 25°C and the curly-winged offspring were scored for their eye colour phenotype on day 15. Asterisks indicate excision frequencies that are significantly greater than the frequency for the Gla mutation at the 5% level.
a Unweighted average frequency of white-eyed flies among curly-winged flies ±se.
In the tests comprising males that did not carry TP5 (left sides of Tables 2 and 3), none of the mutations affected the frequency of SB7 excisions in the germ line. With the ▵2-3 transposase source, these frequencies ranged from 0·31 to 0·42 and the control frequency was 0·34, and with the CP transposase source, they ranged from 0·27 to 0·36 and the control frequency was 0·33. In the presence of a maternally inherited TP5 element, the control SB7 excision frequencies were significantly reduced – to 0·09 in the experiment with the ▵2-3 transposase source and to 0·03 in the experiment with the CP transposase source. Thus, as expected, the maternally inherited TP5 element repressed P-transgene excision in the male germ line. However, this repression was significantly impaired in the flies from the aubQC42, aub▵P−3a and Su(var)2054 mutant stocks in both experiments and in the flies from the piwi1 mutant stock in the experiment with the CP transposase source. By contrast, the flies from the piwi2 mutant stock did not impair repression in either experiment. It is important to note that the mutations tested in these experiments were not present in the males in which the P-transgene excisions occurred; rather, they were present in heterozygous condition in the mothers of these males. Thus, the impairment of repression seen in Tables 2 and 3 was apparently due to dominant maternal effects of the mutations.
To verify that the impairment was caused by the mutations and not by some other factor, we measured CP-induced SB7 excision frequencies in males derived from TP5 w; CyO, SB7/+ stocks from which the various mutations had been removed many generations previously. These mutation-free stocks are expected to have different genetic backgrounds than the stocks that carried the mutations because outcrossing was required to produce them. However, they have the same X chromosome and therefore serve as controls against the possibility that the regulatory function of the TP5 element was altered by random changes in the XL telomere, which is a genetically dynamic structure. The results, shown on the right in Table 3, demonstrate that the removal of the aubQC42, aub▵P−3a and Su(var)2054 mutations from the original stocks restored repression of SB7 excision to a strong level. The impairment of repression that was seen when these mutations were present must therefore be due to the mutations themselves, not to some other factor. By contrast, removal of the piwi1 mutation did not strengthen repression significantly, and removal of the piwi2 mutation actually weakened it. Thus, other factors such as changes in the structure of XL may have influenced the regulatory function of the TP5 element in the piwi mutant stocks or in their mutation-free derivatives.
(iii) Effects of mutations on levels of P-element mRNAs in the female germ line
As the aub and Su(var)205 mutations impair P-element regulation through dominant maternal effects, we used RT–PCR to determine if, in heterozygous condition, these mutations affect the levels of P mRNAs in the maternal germ line; we also tested the piwi mutations for such effects. TP5 w; CyO, SB7/mutation females were crossed to w; CP males to produce the TP5 w/w; CP/mutation females that were used in this analysis. Control females were obtained by crossing w females with w; CP males. RNA was extracted from four samples of each of the various types of females, reverse transcribed into cDNA using an oligo-dT primer, and then the cDNA was amplified by PCR with other primers specific for mRNA from the singed gene (to assess RNA input levels) or mRNAs from the TP5 or CP elements. The results from these experiments, each utilizing the same four sets of cDNA samples, are presented in Figs 1 through 4.
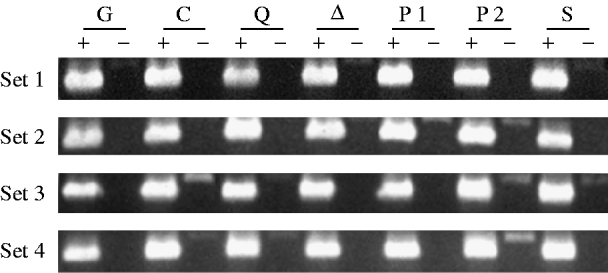
Fig. 1. RT–PCR analysis of singed mRNA from mutant and control female genotypes. Each genotype is represented by four independently obtained samples (sets 1–4). A plus denotes where an aliquot of a sample has been reverse transcribed, and a minus denotes where it has not. The genotypes are TP5 w/w; CP/Gla (lane G), w/w; CP/+ (lane C), TP5 w/w; CP/aubQC42 (lane Q), TP5 w/w; CP/aub▵P−3a (lane ▵), TP5 w/w; CP/piwi1 (lane P1), TP5 w/w; CP/piwi2 (lane P2), and TP5 w/w; CP/Su(var)2054 (lane S). The 630 bp products in the RT(+) lanes were obtained by amplifying the cDNAs with the primers sn-u and sn-d. The 774 bp products seen in some of the RT(−) lanes result from the amplification of contaminating genomic DNA.
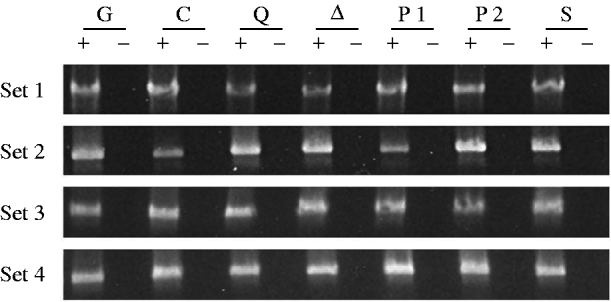
Fig. 2. RT–PCR analysis of somatic CP mRNA from mutant and control female genotypes. The sets of samples and genotypes are as in Fig. 1. The 1539 bp products were obtained by amplifying the cDNAs with the primers P▵0/1-d and P2075-u.

Fig. 3. RT–PCR analysis of germ-line CP mRNA from mutant and control female genotypes. The sets of samples and genotypes are as in Fig. 1. The 1495 bp products were obtained by amplifying the cDNAs with the primers P▵0/1-d and P▵2/3-u.

Fig. 4. RT–PCR analysis of germ-line TP5 mRNA from mutant and control female genotypes. The sets of samples and genotypes are as in Fig. 1. The 471 bp products were obtained by amplifying the cDNAs with the primers TP5-d and P▵2/3-u.
Figure 1 shows the results of amplifying cDNAs derived from RT of mRNAs from the singed gene, which is expressed in the female germ line. The band intensities among the samples within each set are quite similar, indicating that roughly equal amounts of mRNA from the different genotypes were analysed.
Figure 2 shows the results of amplifying cDNAs derived from RT of CP mRNAs that retain the last P intron. Because this intron is not spliced out of P-element transcripts in the somatic cells, we refer to this class of mRNAs as ‘somatic.’ However, it is likely that these mRNAs are also produced in the germ line through incomplete splicing of CP transcripts; see Simmons et al. (Reference Simmons, Haley, Grimes, Raymond and Niemi2002a) for genetic evidence on this point. Figure 2 does not show any consistent differences in the intensities of the PCR products from the various samples that were analysed. The presence of the TP5 element or the presence of an aub, piwi, or Su(var)205 mutation therefore does not seem to affect the level of somatic CP mRNA in females. This finding is consistent with genetic evidence that TP5-mediated system for P regulation does not operate in somatic cells (Stuart et al., Reference Stuart, Haley, Swedzinski, Lockner, Kocian and Simmons2002; Simmons et al., Reference Simmons, Raymond, Niemi, Stuart and Merriman2004).
Figure 3 shows the results of amplifying cDNAs derived from RT of CP mRNAs that have lost the last P intron; these mRNAs are produced exclusively in the germ line. The specificity of this PCR for germ-line CP mRNA is due to the choice of primers, one spanning the first intron, which is in a region deleted in the TP5 element, and one spanning the last intron, which is removed only in germ-line cells. Three observations are noteworthy. First, in three of the four sets of gel images, the PCR product from the TP5 w/w; CP/Gla females (lane G) is clearly fainter than the product from the w/w; CP/+ control females (lane C). This observation confirms previous evidence that the presence of the regulatory TP5 element is associated with a reduction in the level of CP mRNA in the female germ line (Jensen et al., Reference Jensen, Stuart, Goodpaster, Goodman and Simmons2008; Thorp et al., Reference Thorp, Chapman and Simmons2009). Second, in all four sets of gel images, the PCR products from the TP5 w/w; CP/mutation females carrying the aubQC42, aub▵3−Pa or piwi1 alleles are brighter than the products from the TP5 w/w; CP/Gla females (compare lanes Q, ▵ and P1 with lane G). The TP5-mediated loss of CP mRNA is therefore ameliorated in females with these mutant genotypes. Third, the PCR products from the TP5 w/w; CP/mutation females carrying the piwi2 or Su(var)2054 alleles (lanes P2 and S) are fainter than the products from the TP5 w/w; CP/Gla females in two of the four sets of gel images, and they are fainter than the products from the TP5 w/w; CP/mutation females carrying the aubQC42, aub▵P−3a or piwi1 alleles in all four sets of gel images. Thus, little or no amelioration of the loss of CP mRNA occurred in TP5 w/w; CP/piwi2 or TP5 w/w; CP/Su(var)2054 females.
Figure 4 shows the results of amplifying cDNAs derived from RT of TP5 mRNAs in the female germ line. Here, the specificity of the amplification is due to a primer that spans the deletion in the TP5 element; the other primer ensures that only germ-line cDNAs are amplified. TP5 mRNA cannot be produced by the control females (lane C), which lacked a TP5 element. Among the females that carried this element, those heterozygous for the Gla mutation (lane G) yielded little or no PCR product, indicating that they accumulate little TP5 mRNA – an observation that confirms previous evidence that P-element regulation is associated with a paucity of TP5 mRNA (Jensen et al., Reference Jensen, Stuart, Goodpaster, Goodman and Simmons2008; Thorp et al., Reference Thorp, Chapman and Simmons2009). The TP5 w/w females that were heterozygous for the piwi2 mutation also accumulated little of this mRNA. By contrast, the TP5 females that were heterozygous for the aubQC42, aub▵P−3a, piwi1 or Su(var)2054 mutations yielded more of the TP5-specific PCR product in at least three of the four sets of gel images. Thus, in these females, the paucity of TP5 mRNA associated with P-element regulation is ameliorated.
To connect this analysis of P-element RNA with genetic data on regulation by the TP5 element inherited maternally from the various mutant stocks, we measured the frequency of SB7 excisions in the germ lines of the TP5 w; CyO, SB7/CP brothers of the TP5 w/w; CP/mutation females from which the analysed RNA was extracted. The results, summarized in Table 4, demonstrate that the repression of CP-induced SB7 excision was impaired in males from the aubQC42, aub▵P−3a, piwi1 and Su(var)2054 stocks, but not in males from the piwi2 stock, confirming previous findings (Table 3). Thus, the males with impaired P regulation corresponded to the females with elevated levels of germ-line TP5 mRNA. Impaired regulation is not, however, always associated with more expression of CP mRNA. Females heterozygous for three of the mutations – aubQC42, aub▵P−3a, and piwi1 – did have elevated levels of CP mRNA in their germ lines, but females heterozygous for Su(var)2054, which caused the most severe impairment of P regulation in males, did not.
Table 4. Correlated genetic evidence for maternal effects of heterozygous mutations on TP5-mediated repression of CP-induced P transgene excision in the male germ line

The (TP5) w; CyO, SB7/CP males for these tests were obtained from crosses between TP5 w; CyO, SB7/mutation or w; CyO, SB7/+ (control) females and w; CP males at 25°C. Each male was crossed to three Harwich w females at 25°C and their curly-winged offspring were scored for the eye colour phenotype on day 14. The TP5 w; mutation/CP or w; +/CP (control) sisters of these tested males provided RNA for RT-PCR analysis. Asterisks indicate excision frequencies that are significantly greater than the frequency for the Gla mutation at the 5% level.
a Unweighted average frequency of white-eyed flies among curly-winged flies±se.
4. Discussion
TP5-mediated repression of P-element excision in the male germ line has maternal and zygotic components, and both are essential to bring about the repressive state. Males that have inherited a TP5 element paternally do not repress P excision because they lack the maternal component, and males that have inherited cytoplasm but not TP5 maternally from heterozygous TP5/+ mothers do not repress because they lack the zygotic component (Stuart et al., Reference Stuart, Haley, Swedzinski, Lockner, Kocian and Simmons2002; Simmons et al., Reference Simmons, Raymond, Niemi, Stuart and Merriman2004; Thorp et al., Reference Thorp, Chapman and Simmons2009). These facts imply that some aspect of the TP5 function in the maternal germ line is necessary for the zygotic function of TP5 in the germ line of the male offspring.
Do aub, piwi, or Su(var)205 mutations affect the maternal component of P-element regulation in males? Our transgene excision experiments show that this regulation is impaired when the mothers of the males were heterozygous for a mutation in the aub gene. The Aub protein is normally present in the perinuclear cytoplasm of female germ cells, specifically in a region called the nuage (Brennecke et al., Reference Brennecke, Aravin, Stark, Dus, Kellis, Sachidanandam and Hannon2007). This protein appears to play an important role in producing, processing, or transporting piRNAs, including those generated from the locus in the TAS of the XL telomere into which TP5 is inserted (Brennecke et al., Reference Brennecke, Aravin, Stark, Dus, Kellis, Sachidanandam and Hannon2007, Reference Brennecke, Malone, Aravin, Sachidanandam, Stark and Hannon2008). Females that are heterozygous for a mutant aub allele would be expected to have half as much Aub protein as homozygous wild-type females. Our genetic data show that this mutational depletion of Aub protein in the maternal germ line compromises the regulation of P elements in the germ lines of their male offspring. This compromised regulation cannot be due to a zygotic effect of the aub mutations because the males that were tested did not carry these mutations, and it is not likely to be due to some other factor because removal of the aub mutation from the maternal genotype restored regulation fully. Previous studies (Simmons et al., Reference Simmons, Ryzek, Lamour, Goodman, Kummer and Merriman2007) also seem to rule out the possibility that aub mutations engender bona fide genetic changes – for example, lengthening the array of retrotransposons in the telomere of XL – that could influence the regulatory function of a telomeric P element.
Our RT–PCR analyses show that mutational depletion of Aub protein allows the mRNAs from both the TP5 telomeric element and the transposase-encoding CP element to accumulate in female germ-line cells to higher levels than they otherwise would. In one model based on the analysis of small RNAs (Brennecke et al., Reference Brennecke, Aravin, Stark, Dus, Kellis, Sachidanandam and Hannon2007, Reference Brennecke, Malone, Aravin, Sachidanandam, Stark and Hannon2008; Li et al., Reference Li, Vagin, Lee, Xu, Ma, Xi, Seitz, Horwich, Syrzycka, Honda, Kittler, Zapp, Klattenhoff, Schulz, Theurkauf, Weng and Zamore2009; Tushir et al., Reference Tushir, Zamore and Zhang2009), these mRNAs should be processed into piRNAs with a sense sequence orientation through targeted attacks by antisense piRNAs derived from the telomeric P element; these sense piRNAs should, in turn, target antisense transcripts from the telomeric P element to create more antisense piRNAs. With repetition, this alternating, or ping-pong cycle, is expected to generate a large population of sense and antisense P-element piRNAs and, at the same time, to eliminate P mRNAs. The accumulation of TP5 and CP mRNAs in aub−/aub+ females may therefore be taken as an indication that the ping-pong cycle is impaired, resulting, presumably, in a smaller population of piRNAs. Transmission of P-specific piRNAs from mother to offspring through the egg cytoplasm is thought to be responsible for the maternal component of P-element regulation (Brennecke et al., Reference Brennecke, Malone, Aravin, Sachidanandam, Stark and Hannon2008; Jensen et al., Reference Jensen, Stuart, Goodpaster, Goodman and Simmons2008). A reduced maternal endowment of P-element piRNAs is therefore likely why P-transgene excisions are not effectively repressed in the aub +/aub + sons of aub −/aub + females.
The Aub protein has also been implicated in the regulation of other Drosophila transposons (Vagin et al., Reference Vagin, Sigova, Li, Seitz, Gvozdev and Zamore2006). In one detailed study (Chambeyron et al., Reference Chambeyron, Popkova, Payen-Groschêne, Brun, Laouini, Pelisson and Bucheton2008), sense RNAs of the I element, a non-LTR retrotransposon, were found to accumulate in the oocytes of aub −/aub − females, and concomitantly, the piRNAs from this element were found to decrease. This RNA profile was also observed in the F1 hybrid females derived from crosses between inducer (I strain) males and reactive (R strain) females. I elements become active in the germ lines of these females, causing them to be sterile. Maternally transmitted I-element piRNAs appear to be responsible for repressing this sterility in females from the reciprocal cross (I female×R male) and in females from I strains; see also Brennecke et al. (Reference Brennecke, Malone, Aravin, Sachidanandam, Stark and Hannon2008) . In this system, the Aub protein is evidently needed for the production, processing, or transport of these piRNAs, and for the elimination of I-element mRNAs.
The two piwi mutations analysed here had different – and inconsistent – maternal effects on P regulation in the male germ line. Our genetic data indicate that repression of P-transgene excision was impaired in males from the piwi1 stock but not in males from the piwi2 stock in experiments in which CP was the transposase source; however, it was not impaired in either type of male in an experiment in which ▵2-3 was the transposase source. Previous analyses failed to detect a maternal effect of either piwi1 or piwi2 on CP-induced P excisions from the snw allele in the male germ line (Simmons et al., Reference Simmons, Ryzek, Lamour, Goodman, Kummer and Merriman2007). However, a recent study found that piwi1 disrupted TP5-mediated repression of dysgenic sterility in the daughters of females heterozygous for the mutation, although it did not disrupt repression of dysgenesis by another telomeric P element, TP6 (Belinco et al., Reference Belinco, DiPrima, Wolff, Thorp, Buschette and Simmons2009); by contrast, piwi2 had no effect on repression by either telomeric P element. In a related vein, Josse et al. (Reference Josse, Teysset, Toideschini, Sidor, Anxolabéhère and Ronsseray2007) found that females heterozygous for piwi1 or piwi2 had a reduced capacity for a telomere trans-silencing effect (TSE); however, this reduction was seen only when the females were also heterozygous for the Su(var)2054 mutation. All these genetic data suggest that impairment of P-element regulation by a mutation in the piwi gene may depend on the nature of the mutation, the transposase source, the telomeric P element, the genetic assay and the genotype – for example, whether other compromising mutations such as Su(var)2054 are present. It should be noted that although both piwi mutations are due to transgene insertions, the insertion in piwi1 is expected to be more severe. This insertion is in the first coding exon, whereas the insertion in piwi2 is in the fourth coding exon (Cox et al., Reference Cox, Chao, Baker, Chang, Qiao and Lin1998); piwi2 may therefore encode a partially functional polypeptide, which may explain why it has a less severe mutant phenotype (Lin & Spradling, Reference Lin and Spradling1997) and why it does not impair P regulation.
The RT–PCR analyses reported here indicate that both TP5 and CP mRNAs accumulate in the germ lines of females heterozygous for piwi1, but not in the germ lines of females heterozygous for piwi2. The processing of P-element mRNAs into piRNAs therefore appears to be impaired in the piwi1 heterozygotes but not in the piwi2 heterozygotes. This finding parallels the observation that P-element regulation is compromised in the sons of piwi1 heterozygotes but not in the sons of piwi2 heterozygotes. It is not clear what role the Piwi protein plays in the piRNA pathway. Piwi is a nuclear protein that interacts physically with HP1 (Brower-Toland et al., Reference Brower-Toland, Findley, Jiang, Liu, Yin, Dus, Zhou, Elgin and Lin2007). The Piwi protein also binds piRNAs (Brennecke et al., Reference Brennecke, Aravin, Stark, Dus, Kellis, Sachidanandam and Hannon2007). One possibility is that piRNA–Piwi complexes are involved in the transcriptional repression of transposons, including some P elements (Yin & Lin, Reference Yin and Lin2007).
Su(var)205 was the first gene to be implicated in P-element regulation, and several reports have since confirmed its relevance using different assays (Ronsseray et al., Reference Ronsseray, Lehmann and Anxolabéhère1996, Reference Ronsseray, Marin, Lehmann and Anxolabéhère1998; Marin et al., Reference Marin, Lehmann, Nouaud, Izaabel, Anxolabéhère and Ronsseray2000; Haley et al., Reference Haley, Stuart, Raymond, Niemi and Simmons2005; Josse et al., Reference Josse, Teysset, Toideschini, Sidor, Anxolabéhère and Ronsseray2007; Belinco et al., Reference Belinco, DiPrima, Wolff, Thorp, Buschette and Simmons2009). HP1, the protein encoded by this gene, is associated with chromatin, especially heterochromatin (James et al., Reference James, Eissenberg, Craig, Diethrich, Hobson and Elgin1989). In addition to organizing chromatin, HP1 appears to perform a capping function at the ends of chromosomes, including XL (Fanti et al., Reference Fanti, Giovinazzo, Berloco and Pimpinelli1998); it is also present in the telomeric retrotransposon array and in the TAS (Capkova Frydrykova et al., Reference Capkova Frydrykova, Mason and Archer2008). When HP1 is depleted, as in stocks that are heterozygous for a Su(var)205 mutation, the telomeric capping function is compromised and the chromosomes develop elongated retrotransposon arrays (Savitsky et al., Reference Savitsky, Kravchuk, Melnikova and Gvozdev2002). This genetic change may contribute to the impairment of TP5-mediated regulation of P element activity that is observed in such stocks (Haley et al., Reference Haley, Stuart, Raymond, Niemi and Simmons2005).
The RT–PCR experiments indicate that females heterozygous for TP5 and the Su(var)2054 mutation produce more germ-line TP5 mRNA than control females lacking the mutation. However, given the telomere-capping and chromatin-organizing role of HP1, the increase in germ-line TP5 mRNA in females heterozygous for the Su(var)2054 mutation is not likely due to a malfunction in the ping-pong cycle of the piRNA pathway, as it probably is in females heterozygous for an aub mutation. Rather, the increase might stem from more vigorous senseward transcription of the TP5 element. Elongated telomeric retrotransposon arrays of the sort that develop in stocks with Su(var)205 mutations are known to increase the expression of elements inserted in the TAS (Golubovsky et al., Reference Golubovsky, Konev, Walter, Biessmann and Mason2001). This increased expression could result from stimulation of the TP5 promoter by enhancers within the elongated retrotransposon array, or from read-through transcription originating in the retrotransposon array. Alternately, the increased senseward transcription of TP5 in Su(var)2054/+females could simply be due to the depletion of HP1 within the TAS (see Capkova Frydrykova et al., Reference Capkova Frydrykova, Mason and Archer2008).
One indication that the ping-pong cycle of the piRNA pathway is not disrupted by the Su(var)2054 mutation is that the CP mRNA level is low in the germ lines of females heterozygous for this mutation. The RT–PCR analysis (Fig. 3) shows that this mRNA level is consistently lower than the levels in TP5 w/w; CP/mutation females heterozygous for the aubQC42, aub▵P−3a, or piwi1 alleles, which all impaired P regulation, and that it is as low or lower than the level in w/w; CP/+ control females, which cannot carry out the piRNA pathway for P regulation because they lack the critical TP5 element. The comparative dearth of germ-line CP mRNA in TP5 w/w; CP/Su(var)2054 females implies that this mRNA is being processed successfully by the ping-pong cycle to feed the piRNA pathway. Impairment of P regulation by the Su(var)2054 mutation must therefore be due to a breakdown in some other process. There is, by the way, no dearth of somatic CP mRNA in females carrying the Su(var)2054 mutation (Fig. 2). Thus, the reduction in CP mRNA is limited to the germ line, where the proteins thought to be involved in the ping-pong cycle – Aub and Ago3 – are expressed (Brennecke et al., Reference Brennecke, Aravin, Stark, Dus, Kellis, Sachidanandam and Hannon2007, Reference Brennecke, Malone, Aravin, Sachidanandam, Stark and Hannon2008; Li et al., Reference Li, Vagin, Lee, Xu, Ma, Xi, Seitz, Horwich, Syrzycka, Honda, Kittler, Zapp, Klattenhoff, Schulz, Theurkauf, Weng and Zamore2009; Tushir et al., Reference Tushir, Zamore and Zhang2009), and where P excisions are regulated by telomeric P elements such as TP5. Note also that the level of germ-line CP mRNA in females carrying Su(var)2054 is about the same as that in females carrying the piwi2 allele (Fig. 3), which has not been found to impair P regulation by any assay. Thus, the ping-pong cycle seems to be functioning as well in TP5 w/w; CP/Su(var)2054 females, which carry a proven disruptor of P regulation, as in TP5 w/w; CP/piwi2 females, which do not.
If the ping-pong cycle is not impaired by Su(var)2054, then what is the reason for this mutation's profoundly negative effect on P-element regulation? Josse et al. (Reference Josse, Teysset, Toideschini, Sidor, Anxolabéhère and Ronsseray2007) have proposed that the regulation by telomeric elements and transgenes has both post-transcriptional and chromatin-organizing aspects that depend on small RNAs generated from the telomere. According to this proposal, antisense piRNAs derived from a telomeric P element would be the key ingredients in the post-transcriptional aspect of P regulation because by targeting and destroying P mRNAs, they would minimize the synthesis of the P transposase, which is the agent of P-element movement. However, by itself this destruction is apparently not sufficient for effective repression of P-element activity. Transposase-encoding mRNA is at the same low level in TP5 w/w; CP/Su(var)2054 females as it is in TP5 w/w; CP/piwi2 females (Fig. 3); yet, the Su(var)2054 mutation profoundly impairs P regulation through a maternal effect whereas the piwi2 mutation does not. Effective regulation of P-element activity must therefore involve something more than the destruction of transposase-encoding mRNA. Josse et al. (Reference Josse, Teysset, Toideschini, Sidor, Anxolabéhère and Ronsseray2007) suggest that the additional component is chromatin re-organization. Sense piRNAs produced by the destruction of mRNAs may be feed into the ping-pong cycle to generate a population of antisense piRNAs that subsequently associate with proteins such as Piwi to form complexes that bind to P elements throughout the genome. Such complexes may then re-organize the chromatin locally into a state that prevents the P transposase from catalysing P-element movement. Alternately, the piRNA–protein complexes could facilitate the transfer of a pre-existing repressive state from the telomere to other P elements. This process would require ectopic pairing between the telomeric and non-telomeric P elements. In either scenario, HP1 is likely to be involved because it is a known partner of Piwi and it is present in the telomere. The impairment of P regulation that occurs in flies with mutant Su(var)205 mothers could therefore reflect a breakdown in the ability to form piRNA–protein complexes that effectively re-organize chromatin into a repressed state. HP1-like proteins acting in concert with protein complexes that contain small RNAs have also been implicated in chromatin reorganization in Schizosaccharomyces pombe (Verdel et al., Reference Verdel, Jia, Gerber, Sugiyama, Gygi, Grewal and Moazed2004; Grewal, Reference Grewal2010).
How does this hypothesized chromatin re-organization lead to a repression of P-element movement? One possibility is that HP1 binds to complete P elements and blocks, or significantly attenuates, the synthesis of mRNAs encoding the P transposase. In S. pombe, the end result of RNAi/HP1-mediated chromatin reorganization is transcriptional repression. However, our data indicate that the depletion of HP1 does not result in robust synthesis of transposase-encoding P mRNAs, as would be expected in this model; in fact, germ-line CP mRNA levels remain low in females that are heterozygous for an HP1-depleting mutation. This observation suggests that the regulation of P-element movement may involve a subtler mechanism. Instead of quashing P transcription, HP1-binding may simply prevent the P transposase from acting on its substrates – that is, it restricts the action of the P transposase but not that of the RNA polymerase. In this vein, it should be noted that a model of transcriptional repression implies that the loci into which P elements have been inserted run the risk of being silenced, and for a fly carrying 50–80 P elements in its diploid genome, this much of repression might be an unacceptable level of collateral damage.
The model of P regulation that emerges from all these considerations comprises four main steps. First, in the female germ line, piRNAs are generated from a telomeric P element by an as yet unknown mechanism. Second, these piRNAs are amplified by the ping-pong cycle – a process that involves the targeted destruction of P mRNAs, including the mRNA that encodes the P transposase. The Aub and Ago3 proteins carry out this process. Third, the piRNAs created by ping-pong cycling are transmitted to the offspring through the egg cytoplasm. Fourth, some of these piRNAs form complexes with proteins such as Piwi and HP1 to re-organize chromatin into a state that represses P-element movement. Piwi may act as a mediator between the ping-pong cycle and the chromatin re-organizing events by collecting piRNAs from Aub and guiding them to P elements throughout the genome, whereupon HP1 joins Piwi to foster a repressive chromatin state. In this model, HP1 is hypothesized to act downstream of Aub. However, recent analyses of the telomeric TSE have suggested that HP1 may also be involved in the production of piRNAs, possibly by stimulating the expression of their precursors from the TAS (Todeschini et al., Reference Todeschini, Teysset, Delmarre and Ronsseray2010). This chromatin-organizing protein may therefore act upstream as well as downstream of Aub in the system that regulates P elements.
This work was supported by the Department of Genetics, Cell Biology and Development of the University of Minnesota. Benjamin Berres, Eric Chapman and Wojciech Kraszkiewicz helped to develop the P-transgene excision assay used in the genetic analyses.










