Introduction
Five years ago, Dr. David Agus, then Director of the University of Southern California (USC) Center for Applied Molecular Medicine and USC Westside Norris Cancer Center, and co-Director of the USC-National Cancer Institute Physical Sciences in Oncology Center (among many other titles), approached Olympus with a bold idea to collaborate on a project aimed at changing the future of how we treat cancer. It began with a simple conversation sharing a grand vision, backed by support from Larry Ellison, founder of Oracle Corporation, a company providing cloud-based services. Ultimately it led to the formation of the USC-Olympus Partnership in Multiscale Bioimaging under the umbrella of the new Ellison Institute for Transformative Medicine of USC (Figure 1).

Figure 1: The Lawrence J. Ellison Institute for Transformative Medicine of the University of Southern California.
Dr. Agus, a world-renowned cancer doctor, had been thinking about the limitations of current cancer treatment for a long time, writing books, giving TED talks, and forming companies to look at gene and protein markers. He was constantly evangelizing on the topic of taking a broader, more holistic view of cancer. From his years of treating patients and studying the mechanisms of cancer he came to the conclusion that not only were current methods of diagnosing and treating cancer too narrow and antiquated, but the future of treatment had to involve a broad range of information about a patient's life. This not only involved typical pathology and histology, but also genetics, proteomics, and more dynamic modes of imaging living cancer, both in the patient and by taking tumor cells directly from that patient to interrogate in live cell imaging. All of this needed to be mixed in with an understanding of the patient's life.
To pull this off he needed a new collaborative space, bringing together doctors, cancer biologists, computer scientists, physicists, mathematicians, and corporate partners with significant expertise in relevant fields. He tells the story of a breakfast meeting with his friend Larry Ellison. After sharing his vision, Ellison was convinced this was the way of the future, forming the Lawrence J. Ellison Institute for Transformative Medicine of USC with a gift of $200M to build a new center and help support the future of precision medicine.
Agus began to assemble a team (Figure 2). Certainly, with the support of Ellison there were going to be some formidable interactions with a computing powerhouse, but as the vision explicitly stated, there were going to be many needs.

Figure 2: Dr. David Agus (front) and part of the Ellison Institute team.
Olympus, known for its optical capabilities both in the field of microscopy, and as a major medical company, was a natural partner. Reaching out to local Olympus representatives ultimately led Agus to visit Olympus headquarters in Tokyo and the formation of the partnership, which involved explorations of technologies and methods for precision medicine. A mixture of systems was ultimately supplied for digital pathology slide scanning research, 2D and 3D (widefield and confocal) live cell imaging and analysis, as well as optical experimentation, covering a broad range of studies (Figure 3).

Figure 3: USC Ellison Institute research scientist Harish Sura examines human cancer cells in high-resolution 3D while colleague Seungil Kim (background) and Olympus confocal microscopy specialist Shane Andrews (foreground) look on.
One of the clearest applications of microscope technology for precision medicine is in anti-cancer drug screening. This can follow the more canonical methods of imaging and analysis of known cell lines to elucidate mechanistic pathways of cancer action and the response of those pathways to specific anti-cancer compounds. Another provocative method is to use the patient's own cells, rather than cell lines, to see the effects of those anti-cancer compounds on the patient's cancer.
The heterogeneous nature of cancer has been one of the greatest challenges in cancer treatment. There are hundreds of different genes that can be involved, and the total tumor environment—the entire body, not only local tumor microenvironment—can affect the progression of cancer. Therefore, it is important to take multiple methodological approaches for the study of cancer.
Cell Models in the Study of Cancer
One of the new important ways of doing so is to use 3D cellular models. Traditional anti-cancer compound testing has relied for years on growth of cells in essentially 2D monolayers. However, over the last decade, a growing body of literature has led to the strong scientific consensus that cells respond differently in monolayers than they do in 3D structures. This can have a strong effect on the efficacy of anti-cancer compounds, and a large range of scientific models have arisen in response to the need to generate more physiologically relevant biological systems for in vitro study. These include growth of spheroid models and a range of “organs-on-a-chip” to complex interconnected “bodies-on-a-chip” wherein multiple 3D model organ systems are connected. Two-dimensional cell cultures will undoubtedly remain a key method for screening drugs and answering mechanistic pathway questions, but 3D cultures are being widely adopted as a robust and rigorous new way to validate preliminary results in secondary screening in pharmaceutical companies as well as in academia (Figure 4).

Figure 4: Dr. David Agus and Dr. Seungil Kim examine raw confocal data of patient-derived tumor organoids, an important 3D cellular model being studied by the lab.
Among the many projects at the Ellison Institute, the team has been developing and pioneering some methods in the growth and measurement of patient-derived organoids in microplates for anti-cancer compound testing using multiple microscopy techniques. Olympus systems are being used not only for the imaging, but also true 3D high content analysis of these samples in microplate format (Figures 5A and 5B).

Figure 5: A. 3D visualization of patient-derived tumor organoids. B. True 3D object recognition within the same sample. Images courtesy of Ellison Institute.
Tissue Clearing Techniques
One challenge all these models pose with light microscopy imaging is the simple ability to generate clear images. Light scatters as it travels through biological samples, and as these models get thicker and thicker traditional microscopy methods that may have worked well enough in relatively 2D samples suffer from haze and blur in the images, or simple inability to get light deep enough into the sample. Tissue clearing techniques alter the refractive index of samples allowing 3D imaging deep into tissues and organoids (Figures 6A and 6B). Therefore, many scientists have turned to confocal and even 2-photon microscopy to improve the resolution of imaging in these thicker samples. This is especially important when working in the context of live samples.

Figure 6: Confocal imaging of uncleared and cleared organoids showing improved imaging of cells within the organoid. Organoids were approximately 300 µm in diameter. Images were taken in the center of the organoids.
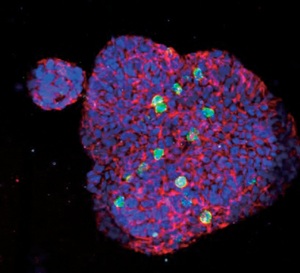
For those choosing to chemically fix samples for endpoint analyses, a wide variety of chemical clearing reagents have been invented by scientists and have been made commercially available. These reagents make otherwise opaque tissue samples transparent through a process of refractive index homogenization [Reference Richardson and Lichtman1]. This is particularly useful when samples have gone through a fluorescent molecular labeling process targeting specific molecules of interest (Figure 7). These labels can then be interrogated by fluorescence microscopy without suffering from the light scattering of the thicker tissue, producing beautiful, spatially resolved images in three dimensions.

Figure 7: Gallery display of objects identified by 3D high content object recognition (center panel), surrounded by more macro-level images of patient-derived colorectal cancer tumor organoids where the objects were found. Nuclei labeled by DAPI in blue, proliferating cells labeled by Ki-67 immunostaining (green), and cell-cell junctions shown with E-cadherin immunostaining (red). Images courtesy of Ellison Institute.
Live Cell and Tissue Imaging
However, such techniques cannot get to an important fourth dimension, time, since the clearing process kills the cells. This is an important factor in cancer research—and any cell biology—since some kinds of mechanistic understanding may only be clearly derived from observations of dynamic interactions over substantially contiguous periods of time. Therefore, microscopy imaging techniques that allow for live cell and tissue interrogation are critical to fill out a more holistic paradigm of precision medicine.
If that paradigm is going to become clinically relevant, there also needs to be further effort towards standardizing the methods and streamlining workflows to make it practical to deploy these technologies in an efficient way, so automation is an important part of the overall project goals to speed up the workflow. Likewise, this can improve reproducibility of results. Certainly, automation of advanced research microscopes is needed, but there are other kinds of standardization that are important.
One of the key teams that Ellison Institute formed was a cell culture team. Early on they recognized that the maintenance and quality control of their cell lines was going to be a critical aspect of their work. Cell lines form the basis of a great deal of scientific research. A perhaps underappreciated fact is that scientific results can be changed quite dramatically if proper care is not taken for consistent growth, passaging, and characterization of cell lines. This can include tracking passage number, confluency, and passage timing, as well as genetic and phenotypic characterization of cell lines, even (and sometimes especially) immortalized cell lines (Figure 8). An increasing body of literature has documented phenotypic changes resulting from different passage number, confluency at time of passage and imaging, as well as misidentification or contamination of immortalized cell lines [2–Reference Trajkovic6]. While proper cell culture quality control is a key element of the work at the Ellison Institute (and also a crucial core facility feature at many major pharmaceutical companies), cell culture is a notoriously labor-intensive process, so the Ellison Institute team has been testing out new remote cell culture monitoring equipment from Olympus. These in-incubator monitoring systems are designed to reduce the time required to constantly check cell cultures for confluency, while automatically documenting growth curves using machine learning (ML) applied to images of the cell culture. With proper IT conditions, the Ellison Institute team has been able to monitor their cultures from off-campus, something that may continue to be useful in a post-COVID-19 world. And for anyone regularly conducting cell-based assays, the NIH's National Center for Advancement of Translational Sciences Assay Guidance Manual clearly recommends rigorous monitoring of cell culture passage number and confluency among other controls (Figures 8 and 9).

Figure 8: Comparison display of two different growth curves during successive passages. A change in the cell growth curve indicates that further culture of the cells should be discontinued. Such automated comparative data can be used to check cell culture quality and consistency.

Figure 9: The typical cell culture process involves many manual and qualitative assessments of cell culture quality and confluency. Automation of that process can lead to more consistent assay.
The Role of Artificial Intelligence
In the view of Dr. Agus and the Ellison Institute team, it will take a range of technologies, including some new abilities, in order to effectively advance precision cancer medicine, but it is not only the optical technologies that are important in this effort. In the past several decades, combined advances in computing power, algorithmic analysis methods, deep data mining, and especially artificial intelligence (AI) have opened up vast new capacity to not only analyze individual data sets, but to provide cross-correlation of disparate data elements in the entire patient journey.
AI is already being investigated in many parts of that journey, from analysis of patient records to genetic databases. Already deployed are a range of ML and AI methods for analysis of tissue and cellular imaging. ML and AI have shown clear utility in analysis of histopathology and toxicology of sectioned tissues, as well as analysis of statistically significant cell populations as is commonly done in high content analysis (HCA) (Figures 10 and 11). These kinds of analyses can help to identify different cell types, organ and tissue type boundaries, cancerous versus normal tissue, and subcellular events such as nuclear translocation. Much of this is aided by the advancing field of biomarker research, which can specifically label cancer-associated molecules.

Figure 10: Scatter plot display of object recognition linked to image of identified object. Such object recognition can be enhanced by AI methods during the process. Image data courtesy of Ellison Institute.

Figure 11: Staurosporine (STS) induced HT-29 human colorectal cancer cell apoptosis measured in 3D spheroids indicating increased apoptosis with increasing concentration of STS. Nuclei labeled by Hoechst 33342 (blue); apoptotic cell nuclei labeled with NucView 550 (magenta).
The power of ML/AI, especially deep learning AI, opens up the potential for training of computer algorithms to automatically identify different cell populations, despite variations in image shading or other irregularities which the trained human eye may be able to adjust for but until recently was very difficult for computers. More powerfully, trained AI can easily recognize specific features and provide quantitative measurements in a fraction of the time that it would take a human doing manual measurements. While the field is advancing and further research is required to improve methods, ML/AI has undoubtedly advanced the field of analysis, and Olympus is pushing forward with this technology and testing it in the collaboration with the Ellison Institute.
Not only does this provide the ability to speed up analysis, but it also has the potential to identify new patterns in biology that might otherwise be too subtle or complex to distinguish by eye. In the field of cancer biology, particularly with all the cancer biomarkers being identified, analysis of the expression levels and interactions of these markers becomes ever more challenging, and the application of sophisticated AI in this context will help untangle the complexity.
The Future
Despite the challenges of the current pandemic, the Ellison Institute recently opened its new building, which has both scientific and clinical spaces. The passion of the team and their collaborative spirit shows great promise. Multiple efforts are now underway globally working towards Precision Medicine, including the recent Lifetime Initiative, a 10-year, one-hundred-billion-dollar European initiative to advance a broad range of collaborative studies across multiple fields with a similar philosophy pioneered by Dr. Agus and Larry Ellison. Clearly the vision was right, and so the work continues towards a more integrated and intelligent future for medicine.