Published online by Cambridge University Press: 08 March 2022
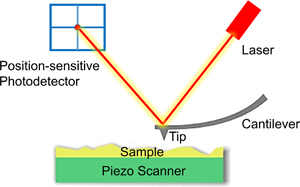
Tumors have posed a serious threat to human life and health. Researchers can determine whether or not cells are cancerous, whether the cancer cells are invasive or metastatic, and what the effects of drugs are on cancer cells by the physical properties such as hardness, adhesion, and Young's modulus. The atomic force microscope (AFM) has emerged as a key important tool for biomechanics research on tumor cells due to its ability to image and collect force spectroscopy information of biological samples with nano-level spatial resolution and under near-physiological conditions. This article reviews the existing results of the study of cancer cells with AFM. The main foci are the operating principle of AFM and research advances in mechanical property measurement, ultra-microtopography, and molecular recognition of tumor cells, which allows us to outline what we do know it in a systematic way and to summarize and to discuss future directions.