Article contents
Towards Augmented Microscopy with Reinforcement Learning-Enhanced Workflows
Published online by Cambridge University Press: 05 September 2022
Abstract
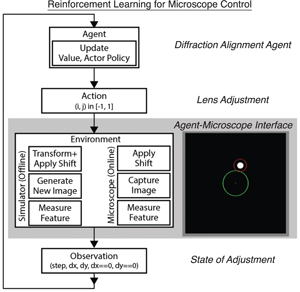
Here, we report a case study implementation of reinforcement learning (RL) to automate operations in the scanning transmission electron microscopy workflow. To do so, we design a virtual, prototypical RL environment to test and develop a network to autonomously align the electron beam position without prior knowledge. Using this simulator, we evaluate the impact of environment design and algorithm hyperparameters on alignment accuracy and learning convergence, showing robust convergence across a wide hyperparameter space. Additionally, we deploy a successful model on the microscope to validate the approach and demonstrate the value of designing appropriate virtual environments. Consistent with simulated results, the on-microscope RL model achieves convergence to the goal alignment after minimal training. Overall, the results highlight that by taking advantage of RL, microscope operations can be automated without the need for extensive algorithm design, taking another step toward augmenting electron microscopy with machine learning methods.
- Type
- Software and Instrumentation
- Information
- Copyright
- Copyright © The Author(s), 2022. Published by Cambridge University Press on behalf of the Microscopy Society of America
References
- 10
- Cited by



